Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN751-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN751-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 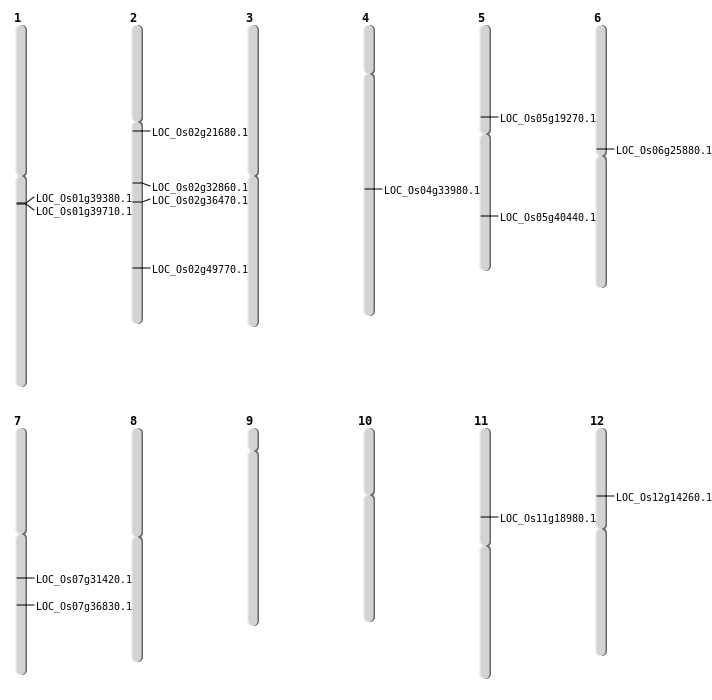
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1881445 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 5593603 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 12557479 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 15136578 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 23044567 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 3575931 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 13326191 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 13176929 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 21186410 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 12886235 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g21680 |
| SBS | Chr2 | 27913978 | A-C | HOMO | INTRON | |
| SBS | Chr2 | 9573138 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 29847884 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 628677 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 628679 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10003955 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 20175754 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 20570938 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g33980 |
| SBS | Chr4 | 2884068 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 2884182 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 3081352 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 3291499 | A-G | HET | INTRON | |
| SBS | Chr5 | 11048269 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 20259236 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 23757604 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g40440 |
| SBS | Chr6 | 13740531 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 15121095 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g25880 |
| SBS | Chr6 | 17773913 | T-C | HET | INTRON | |
| SBS | Chr7 | 22150071 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 22150072 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 15084559 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 27369671 | C-G | HOMO | INTRON | |
| SBS | Chr9 | 11864800 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 8615368 | T-G | HOMO | INTRON |
Deletions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 6491681 | 6491682 | 1 | |
| Deletion | Chr12 | 8120121 | 8120126 | 5 | LOC_Os12g14260 |
| Deletion | Chr7 | 18611730 | 18611731 | 1 | LOC_Os07g31420 |
| Deletion | Chr2 | 19513539 | 19513545 | 6 | LOC_Os02g32860 |
| Deletion | Chr4 | 22048118 | 22048120 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 5167946 | 5167947 | 2 | |
| Insertion | Chr6 | 20611402 | 20611403 | 2 | |
| Insertion | Chr6 | 575571 | 575572 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 8134302 | 8392133 | |
| Inversion | Chr11 | 10814852 | 10816458 | LOC_Os11g18980 |
| Inversion | Chr2 | 22030258 | 22528744 | LOC_Os02g36470 |
| Inversion | Chr1 | 22166460 | 22396559 | 2 |
| Inversion | Chr2 | 22528752 | 30427749 | LOC_Os02g49770 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 7364213 | Chr1 | 22167474 | LOC_Os01g39380 |
| Translocation | Chr5 | 7364223 | Chr1 | 22166813 | LOC_Os01g39380 |
| Translocation | Chr11 | 7874187 | Chr1 | 27872078 | |
| Translocation | Chr5 | 11191552 | Chr2 | 30426493 | LOC_Os05g19270 |
| Translocation | Chr5 | 20408123 | Chr2 | 18029417 | |
| Translocation | Chr7 | 22069284 | Chr6 | 25276920 | LOC_Os07g36830 |
| Translocation | Chr12 | 24683709 | Chr2 | 5772737 | |
| Translocation | Chr6 | 28071057 | Chr1 | 22168539 | LOC_Os01g39380 |
| Translocation | Chr6 | 28074491 | Chr1 | 22396573 | LOC_Os01g39710 |
