Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN764-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN764-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 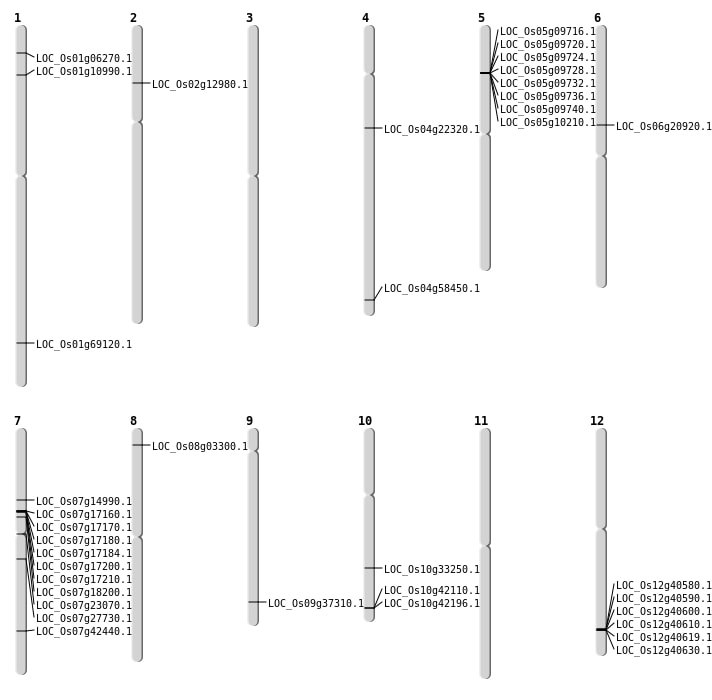
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23498957 | C-A | HET | INTRON | |
| SBS | Chr1 | 5861971 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g10990 |
| SBS | Chr1 | 5904841 | C-A | HET | INTRON | |
| SBS | Chr10 | 15033310 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 17451797 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g33250 |
| SBS | Chr10 | 9316889 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 11448769 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 6277561 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 25172760 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 25538981 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 25540917 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 16146175 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 21201462 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 28984926 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 12634430 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22320 |
| SBS | Chr4 | 13593744 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 14547660 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 28580209 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 34773312 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g58450 |
| SBS | Chr5 | 3579783 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 4722171 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 7165376 | A-T | HET | INTRON | |
| SBS | Chr6 | 13665621 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 4158956 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 10092053 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g17160 |
| SBS | Chr7 | 13013367 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g23070 |
| SBS | Chr7 | 153340 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 16179008 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g27730 |
| SBS | Chr8 | 22613818 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 23577137 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 542779 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 7642844 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 14090251 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 21553825 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g37310 |
| SBS | Chr9 | 6179294 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 9717118 | C-T | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2184012 | 2184013 | 1 | |
| Deletion | Chr9 | 3852373 | 3852385 | 12 | |
| Deletion | Chr5 | 5495001 | 5555000 | 60000 | 8 |
| Deletion | Chr4 | 6695116 | 6695123 | 7 | |
| Deletion | Chr2 | 6882041 | 6882059 | 18 | LOC_Os02g12980 |
| Deletion | Chr2 | 7004524 | 7004544 | 20 | |
| Deletion | Chr1 | 7244354 | 7244355 | 1 | |
| Deletion | Chr10 | 8698506 | 8698510 | 4 | |
| Deletion | Chr7 | 10093001 | 10140000 | 47000 | 6 |
| Deletion | Chr6 | 12082660 | 12082662 | 2 | LOC_Os06g20920 |
| Deletion | Chr4 | 12634734 | 12634741 | 7 | |
| Deletion | Chr1 | 13622575 | 13622581 | 6 | |
| Deletion | Chr6 | 14723302 | 14723317 | 15 | |
| Deletion | Chr10 | 18094826 | 18094827 | 1 | |
| Deletion | Chr5 | 20364542 | 20364562 | 20 | |
| Deletion | Chr10 | 22652636 | 22652637 | 1 | LOC_Os10g42110 |
| Deletion | Chr10 | 22844975 | 22845003 | 28 | |
| Deletion | Chr11 | 22938450 | 22938465 | 15 | |
| Deletion | Chr12 | 25114001 | 25169000 | 55000 | 6 |
| Deletion | Chr2 | 25212280 | 25212358 | 78 | |
| Deletion | Chr7 | 25410263 | 25410266 | 3 | LOC_Os07g42440 |
| Deletion | Chr3 | 35917472 | 35917484 | 12 | |
| Deletion | Chr1 | 40178547 | 40178562 | 15 | LOC_Os01g69120 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2329880 | 2329881 | 2 | |
| Insertion | Chr1 | 5726034 | 5726035 | 2 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 1532791 | 2152604 | LOC_Os08g03300 |
| Inversion | Chr8 | 8352011 | 8975028 | |
| Inversion | Chr8 | 8352033 | 8975021 | |
| Inversion | Chr7 | 8597714 | 10093460 | LOC_Os07g14990 |
| Inversion | Chr7 | 10790798 | 10926322 | LOC_Os07g18200 |
| Inversion | Chr7 | 10920496 | 10926327 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 1532808 | Chr1 | 2975375 | 2 |
| Translocation | Chr8 | 2039401 | Chr1 | 2975388 | LOC_Os01g06270 |
| Translocation | Chr10 | 22697606 | Chr6 | 4198232 | |
| Translocation | Chr10 | 22699236 | Chr6 | 14677084 | LOC_Os10g42196 |
