Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN769-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN769-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 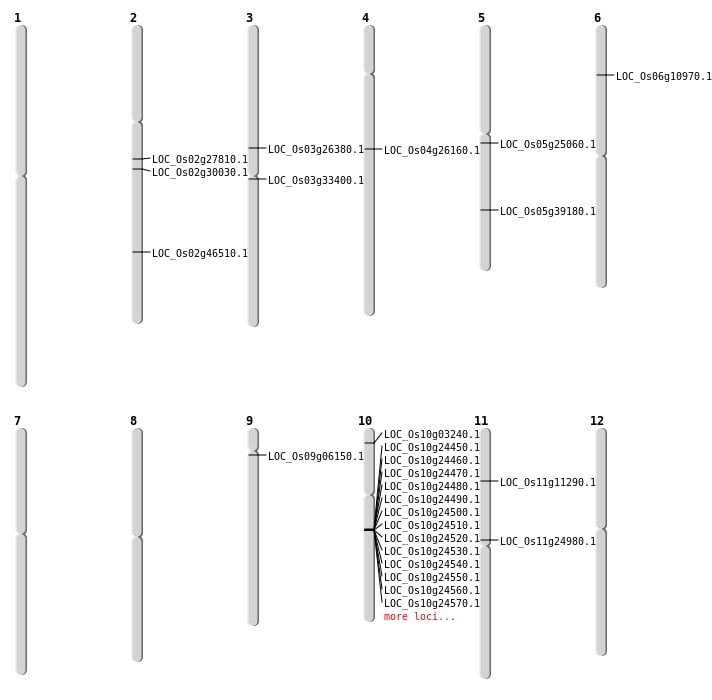
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22473862 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr1 | 39194785 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 42767777 | A-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 9707213 | G-T | HET | INTRON | |
| SBS | Chr10 | 12924228 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 9765241 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 12724547 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 22608758 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 2350784 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 6251849 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g11290 |
| SBS | Chr12 | 21887347 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 377180 | T-C | HET | INTRON | |
| SBS | Chr2 | 13610472 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 14181964 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 17208586 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 29497971 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 15080411 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g26380 |
| SBS | Chr3 | 19097989 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g33400 |
| SBS | Chr4 | 12533306 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 15258737 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g26160 |
| SBS | Chr5 | 14500998 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g25060 |
| SBS | Chr5 | 15366083 | C-T | HET | INTRON | |
| SBS | Chr5 | 19262218 | T-G | HET | INTRON | |
| SBS | Chr5 | 23799208 | C-T | HET | INTRON | |
| SBS | Chr7 | 11617090 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 173274 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 24399436 | C-T | HET | INTRON | |
| SBS | Chr7 | 8638678 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 9201009 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 2576489 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 11977751 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr9 | 13212553 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 1606559 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 2879465 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g06150 |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2711685 | 2711687 | 2 | |
| Deletion | Chr6 | 3574791 | 3574793 | 2 | |
| Deletion | Chr12 | 4229748 | 4229749 | 1 | |
| Deletion | Chr6 | 5732846 | 5732850 | 4 | LOC_Os06g10970 |
| Deletion | Chr4 | 6528438 | 6528459 | 21 | |
| Deletion | Chr9 | 7472473 | 7472548 | 75 | |
| Deletion | Chr1 | 10808907 | 10808908 | 1 | |
| Deletion | Chr5 | 12074056 | 12074060 | 4 | |
| Deletion | Chr10 | 12545001 | 12663000 | 118000 | 20 |
| Deletion | Chr11 | 14224097 | 14224104 | 7 | LOC_Os11g24980 |
| Deletion | Chr1 | 14480258 | 14480267 | 9 | |
| Deletion | Chr1 | 15176174 | 15176183 | 9 | |
| Deletion | Chr2 | 17841456 | 17841458 | 2 | LOC_Os02g30030 |
| Deletion | Chr12 | 18804445 | 18804447 | 2 | |
| Deletion | Chr2 | 19820358 | 19820366 | 8 | |
| Deletion | Chr3 | 21067983 | 21067999 | 16 | |
| Deletion | Chr3 | 22003256 | 22003268 | 12 | |
| Deletion | Chr3 | 23613828 | 23613843 | 15 | |
| Deletion | Chr5 | 23795454 | 23795456 | 2 | |
| Deletion | Chr8 | 27156353 | 27156371 | 18 | |
| Deletion | Chr8 | 27663722 | 27663784 | 62 | |
| Deletion | Chr5 | 27977435 | 27977437 | 2 | |
| Deletion | Chr2 | 28355196 | 28356197 | 1001 | LOC_Os02g46510 |
| Deletion | Chr2 | 30741076 | 30741077 | 1 | |
| Deletion | Chr1 | 32042630 | 32042631 | 1 | |
| Deletion | Chr2 | 33185688 | 33187437 | 1749 | |
| Deletion | Chr1 | 38322409 | 38322415 | 6 | |
| Deletion | Chr1 | 42478357 | 42478516 | 159 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 16468454 | 16468457 | 4 | LOC_Os02g27810 |
| Insertion | Chr2 | 18588040 | 18588050 | 11 | |
| Insertion | Chr2 | 19427799 | 19427804 | 6 | |
| Insertion | Chr4 | 25055851 | 25055851 | 1 | |
| Insertion | Chr5 | 21200975 | 21200976 | 2 | |
| Insertion | Chr7 | 16456453 | 16456453 | 1 | |
| Insertion | Chr7 | 28354245 | 28354246 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 1031958 | 1370352 | LOC_Os10g03240 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1030941 | Chr4 | 16124871 | |
| Translocation | Chr5 | 22977252 | Chr2 | 28355198 | LOC_Os05g39180 |
