Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN778-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN778-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 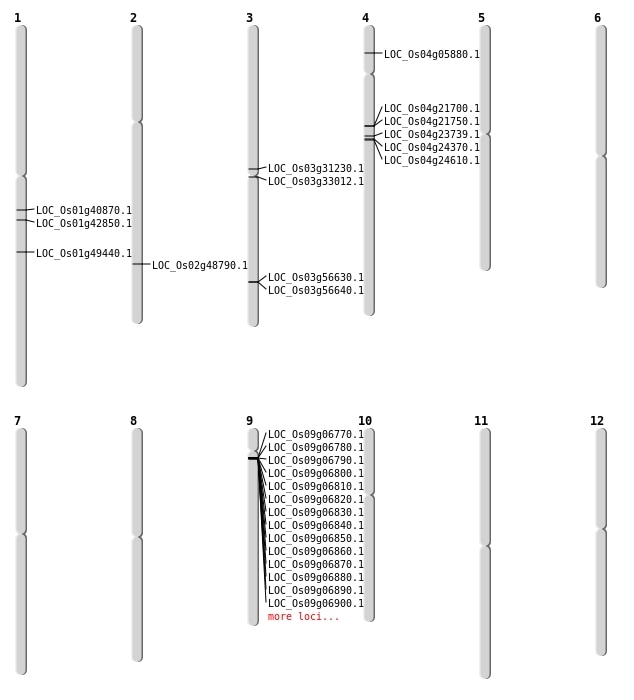
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12165144 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 23118999 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g40870 |
| SBS | Chr1 | 27519110 | A-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 10510231 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 12407415 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 5875812 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 11179822 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 18616749 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 20263978 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 23858455 | G-C | HET | INTRON | |
| SBS | Chr12 | 10998902 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 22628590 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 16686961 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 2106300 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 26392004 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 29864978 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g48790 |
| SBS | Chr3 | 31077730 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 31077731 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 31077732 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 13573227 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g23739 |
| SBS | Chr4 | 13977688 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g24370 |
| SBS | Chr4 | 27742565 | T-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 3026352 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05880 |
| SBS | Chr4 | 33688783 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 33688784 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 5519135 | A-T | HET | INTRON | |
| SBS | Chr4 | 5519138 | A-T | HET | INTRON | |
| SBS | Chr5 | 12302216 | G-A | HET | INTRON | |
| SBS | Chr6 | 17083918 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 28049251 | A-G | HET | INTRON | |
| SBS | Chr7 | 11640255 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 1528106 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 18226425 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 23951939 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 22016696 | T-A | HOMO | INTRON | |
| SBS | Chr8 | 27354742 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 18595920 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 21237987 | G-A | HET | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 556953 | 556956 | 3 | |
| Deletion | Chr6 | 1493803 | 1493827 | 24 | |
| Deletion | Chr1 | 2649533 | 2649581 | 48 | |
| Deletion | Chr9 | 3236001 | 3285000 | 49000 | 9 |
| Deletion | Chr9 | 3287001 | 3328000 | 41000 | 6 |
| Deletion | Chr8 | 5072041 | 5072042 | 1 | |
| Deletion | Chr11 | 5679205 | 5679211 | 6 | |
| Deletion | Chr2 | 10756423 | 10756427 | 4 | |
| Deletion | Chr5 | 11402625 | 11402631 | 6 | |
| Deletion | Chr4 | 13573205 | 13573215 | 10 | LOC_Os04g23739 |
| Deletion | Chr4 | 16387033 | 16387037 | 4 | |
| Deletion | Chr4 | 17334294 | 17334296 | 2 | |
| Deletion | Chr7 | 20974135 | 20974136 | 1 | |
| Deletion | Chr6 | 21184356 | 21184372 | 16 | |
| Deletion | Chr12 | 23473973 | 23473976 | 3 | |
| Deletion | Chr5 | 23596554 | 23596563 | 9 | |
| Deletion | Chr12 | 24953500 | 24953501 | 1 | |
| Deletion | Chr11 | 26328377 | 26328379 | 2 | |
| Deletion | Chr8 | 27294064 | 27294070 | 6 | |
| Deletion | Chr1 | 28433219 | 28433220 | 1 | LOC_Os01g49440 |
| Deletion | Chr2 | 29011645 | 29011655 | 10 | |
| Deletion | Chr5 | 29545083 | 29545089 | 6 | |
| Deletion | Chr2 | 29866565 | 29866573 | 8 | LOC_Os02g48790 |
| Deletion | Chr3 | 30129181 | 30129383 | 202 | |
| Deletion | Chr2 | 30520234 | 30520238 | 4 | |
| Deletion | Chr3 | 32254326 | 32261250 | 6924 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 25252911 | 25252911 | 1 | |
| Insertion | Chr12 | 23551114 | 23551115 | 2 | |
| Insertion | Chr4 | 7291290 | 7291290 | 1 |
Inversions: 5
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 3165873 | Chr1 | 24455555 | |
| Translocation | Chr2 | 3165879 | Chr1 | 24386184 | LOC_Os01g42850 |
| Translocation | Chr7 | 23893139 | Chr3 | 18885718 | LOC_Os03g33012 |
| Translocation | Chr7 | 23893149 | Chr3 | 17781360 | LOC_Os03g31230 |
