Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN791-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN791-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 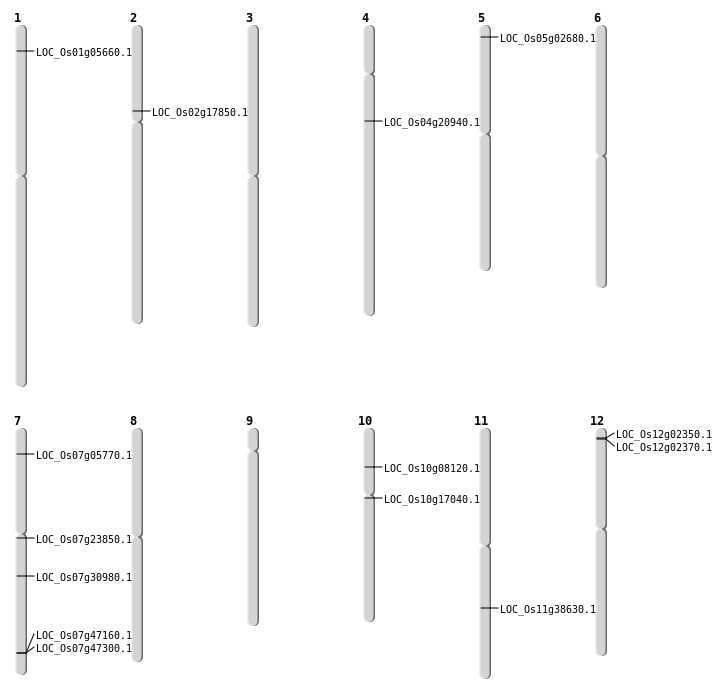
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17039902 | C-A | HET | INTRON | |
| SBS | Chr1 | 26328581 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 29344350 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 3393632 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 39892799 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 587755 | A-T | HET | UTR_5_PRIME | |
| SBS | Chr10 | 21903614 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 4390154 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g08120 |
| SBS | Chr12 | 14011849 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 768579 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 768580 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 32972612 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 33031181 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 16982770 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 30831750 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 12881415 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 12881416 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 14642371 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 21182393 | G-A | HET | INTRON | |
| SBS | Chr5 | 24448648 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 6694398 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 951238 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g02680 |
| SBS | Chr5 | 951241 | G-T | HET | STOP_GAINED | LOC_Os05g02680 |
| SBS | Chr6 | 29594936 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 30436203 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 446464 | G-C | HOMO | INTRON | |
| SBS | Chr6 | 644525 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 16788362 | T-C | HET | INTRON | |
| SBS | Chr7 | 19404910 | G-T | HOMO | UTR_3_PRIME | |
| SBS | Chr7 | 20783800 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 28278051 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g47300 |
| SBS | Chr7 | 28278052 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g47300 |
| SBS | Chr8 | 13777220 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 28026373 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 5496117 | A-C | HET | INTERGENIC |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 764405 | 768522 | 4117 | 2 |
| Deletion | Chr7 | 2781039 | 2781042 | 3 | LOC_Os07g05770 |
| Deletion | Chr4 | 4385672 | 4385678 | 6 | |
| Deletion | Chr7 | 7051152 | 7051153 | 1 | |
| Deletion | Chr10 | 8006794 | 8006799 | 5 | |
| Deletion | Chr2 | 10333506 | 10333530 | 24 | LOC_Os02g17850 |
| Deletion | Chr1 | 12184186 | 12184200 | 14 | |
| Deletion | Chr7 | 13471192 | 13471200 | 8 | LOC_Os07g23850 |
| Deletion | Chr6 | 17140649 | 17140656 | 7 | |
| Deletion | Chr7 | 18348090 | 18348096 | 6 | LOC_Os07g30980 |
| Deletion | Chr8 | 20711104 | 20711111 | 7 | |
| Deletion | Chr5 | 22623578 | 22623607 | 29 | |
| Deletion | Chr11 | 22901713 | 22901716 | 3 | LOC_Os11g38630 |
| Deletion | Chr1 | 25984334 | 25984336 | 2 | |
| Deletion | Chr7 | 28193995 | 28194001 | 6 | LOC_Os07g47160 |
| Deletion | Chr3 | 31712761 | 31712762 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 8535827 | 8535828 | 2 | LOC_Os10g17040 |
| Insertion | Chr11 | 22223380 | 22223383 | 4 | |
| Insertion | Chr4 | 3952441 | 3952445 | 5 | |
| Insertion | Chr8 | 13657406 | 13657407 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 11519477 | 11752959 | LOC_Os04g20940 |
| Inversion | Chr4 | 11519493 | 11752963 | LOC_Os04g20940 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 1053644 | Chr1 | 2701864 | LOC_Os01g05660 |
