Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN835-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN835-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 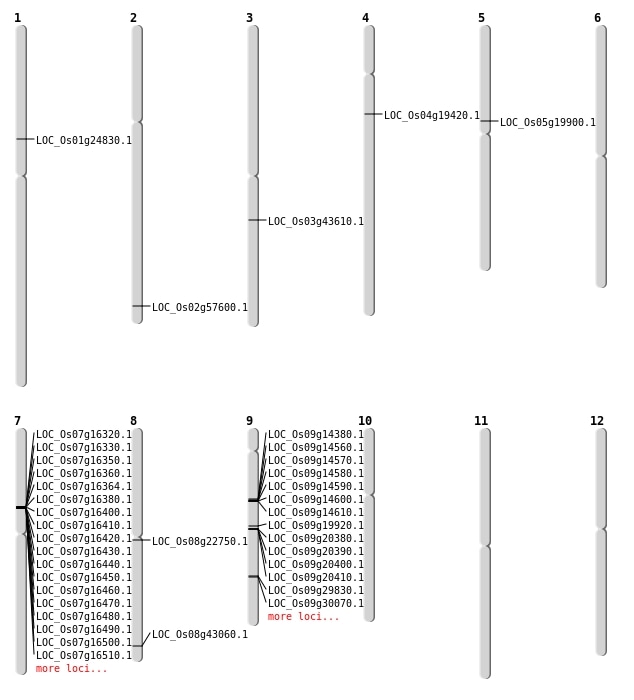
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10266703 | C-T | HET | INTRON | |
| SBS | Chr1 | 13985196 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g24830 |
| SBS | Chr1 | 25080822 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 28886094 | G-C | HET | INTRON | |
| SBS | Chr1 | 30809162 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 31457913 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 9780798 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 16761580 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 3192086 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 7935575 | T-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 12643181 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 14905592 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 18680744 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 25722508 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 12046463 | C-T | HET | INTRON | |
| SBS | Chr2 | 16690944 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 17779826 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 24374368 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g43610 |
| SBS | Chr3 | 34565099 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10820887 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g19420 |
| SBS | Chr4 | 14537431 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 19645806 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 9755681 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 11601972 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g19900 |
| SBS | Chr5 | 1306373 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 16398336 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 19144926 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 27766479 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 3117597 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 3649030 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 17066991 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 8112368 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 12027948 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 18230854 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 27212142 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g43060 |
| SBS | Chr8 | 6115934 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 19737819 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g33480 |
| SBS | Chr9 | 4648015 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 5897478 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 5994745 | T-G | HOMO | INTRON | |
| SBS | Chr9 | 8494703 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14380 |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 969107 | 969119 | 12 | |
| Deletion | Chr10 | 1165337 | 1165340 | 3 | |
| Deletion | Chr6 | 2635694 | 2635698 | 4 | |
| Deletion | Chr4 | 3422418 | 3422419 | 1 | |
| Deletion | Chr8 | 4903508 | 4903509 | 1 | |
| Deletion | Chr3 | 5282317 | 5282319 | 2 | |
| Deletion | Chr8 | 5719648 | 5719657 | 9 | |
| Deletion | Chr8 | 5753475 | 5753489 | 14 | |
| Deletion | Chr11 | 7327246 | 7327257 | 11 | |
| Deletion | Chr9 | 8625001 | 8665000 | 40000 | 6 |
| Deletion | Chr7 | 9530001 | 9710000 | 180000 | 23 |
| Deletion | Chr7 | 10746417 | 10746424 | 7 | |
| Deletion | Chr9 | 11917331 | 11917337 | 6 | LOC_Os09g19920 |
| Deletion | Chr9 | 12232001 | 12282000 | 50000 | 4 |
| Deletion | Chr11 | 13353866 | 13353871 | 5 | |
| Deletion | Chr8 | 13679065 | 13679068 | 3 | LOC_Os08g22750 |
| Deletion | Chr3 | 14081747 | 14081751 | 4 | |
| Deletion | Chr11 | 16210606 | 16210658 | 52 | |
| Deletion | Chr11 | 17132852 | 17132862 | 10 | |
| Deletion | Chr3 | 17593408 | 17593411 | 3 | |
| Deletion | Chr11 | 24900677 | 24900680 | 3 | |
| Deletion | Chr4 | 25305623 | 25305627 | 4 | |
| Deletion | Chr7 | 25393351 | 25393425 | 74 | LOC_Os07g42420 |
| Deletion | Chr1 | 26658400 | 26658416 | 16 | |
| Deletion | Chr3 | 30526747 | 30526763 | 16 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 5201898 | 5201899 | 2 | |
| Insertion | Chr2 | 35278281 | 35278282 | 2 | LOC_Os02g57600 |
| Insertion | Chr5 | 3542560 | 3542560 | 1 | |
| Insertion | Chr7 | 28173836 | 28173836 | 1 | |
| Insertion | Chr8 | 23008681 | 23008681 | 1 | |
| Insertion | Chr9 | 5009001 | 5009004 | 4 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 7908327 | 7997459 | |
| Inversion | Chr9 | 8507407 | 8627632 | |
| Inversion | Chr9 | 18143750 | 18283657 | 2 |
| Inversion | Chr9 | 18283663 | 18317259 | 2 |
No Translocation
