Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN837-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN837-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 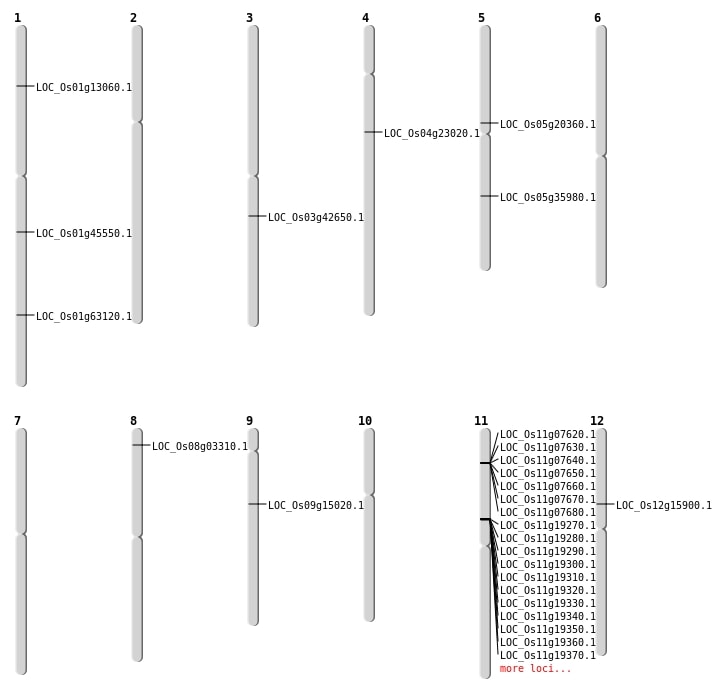
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18013954 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 23648659 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 28566540 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 36580311 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 36580312 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g63120 |
| SBS | Chr1 | 40685653 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 6952290 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 20223846 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 2072352 | T-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 215274 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 27330933 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 3446751 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14554936 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 19509480 | G-T | HOMO | INTRON | |
| SBS | Chr12 | 8954890 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 27432863 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 14681147 | G-A | HET | INTRON | |
| SBS | Chr3 | 25417712 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 29945898 | T-C | HET | INTRON | |
| SBS | Chr4 | 10792584 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 12122074 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 25367743 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 11937331 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20360 |
| SBS | Chr5 | 29010361 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 18092437 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 4248135 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 14026266 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 17213184 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19967134 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 8655409 | G-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 11764175 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 6613034 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 9056079 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g15020 |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 609741 | 609742 | 1 | |
| Deletion | Chr11 | 3874950 | 3921361 | 46411 | 7 |
| Deletion | Chr5 | 4159756 | 4159757 | 1 | |
| Deletion | Chr9 | 5791860 | 5791861 | 1 | |
| Deletion | Chr2 | 6624230 | 6624236 | 6 | |
| Deletion | Chr3 | 6749035 | 6749045 | 10 | |
| Deletion | Chr11 | 8089654 | 8089655 | 1 | |
| Deletion | Chr11 | 10569852 | 10569853 | 1 | |
| Deletion | Chr11 | 11032918 | 11386448 | 353530 | 51 |
| Deletion | Chr3 | 14622955 | 14622956 | 1 | |
| Deletion | Chr9 | 17260174 | 17260183 | 9 | |
| Deletion | Chr10 | 19880077 | 19880078 | 1 | |
| Deletion | Chr4 | 20574010 | 20574011 | 1 | |
| Deletion | Chr1 | 20582029 | 20582046 | 17 | |
| Deletion | Chr4 | 21096219 | 21096223 | 4 | |
| Deletion | Chr5 | 21320342 | 21320343 | 1 | LOC_Os05g35980 |
| Deletion | Chr2 | 24189782 | 24189803 | 21 | |
| Deletion | Chr12 | 24390133 | 24390135 | 2 | |
| Deletion | Chr12 | 25476545 | 25476548 | 3 | |
| Deletion | Chr1 | 25864459 | 25864472 | 13 | LOC_Os01g45550 |
| Deletion | Chr1 | 38082357 | 38082358 | 1 |
Insertions: 7
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 968689 | 1539030 | LOC_Os08g03310 |
| Inversion | Chr8 | 968710 | 1539032 | LOC_Os08g03310 |
| Inversion | Chr1 | 6801628 | 7272843 | LOC_Os01g13060 |
| Inversion | Chr1 | 9838063 | 22691080 | |
| Inversion | Chr1 | 19823071 | 22330331 | |
| Inversion | Chr3 | 23743868 | 25836575 | LOC_Os03g42650 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3874948 | Chr1 | 9837983 | |
| Translocation | Chr11 | 3875234 | Chr1 | 22690998 | |
| Translocation | Chr11 | 3875243 | Chr1 | 6801635 | |
| Translocation | Chr11 | 3921327 | Chr1 | 7272847 | LOC_Os01g13060 |
| Translocation | Chr5 | 7502539 | Chr3 | 16768850 | |
| Translocation | Chr12 | 9052713 | Chr4 | 13069486 | 2 |
| Translocation | Chr8 | 15012535 | Chr7 | 26841517 |
