Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN846-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN846-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 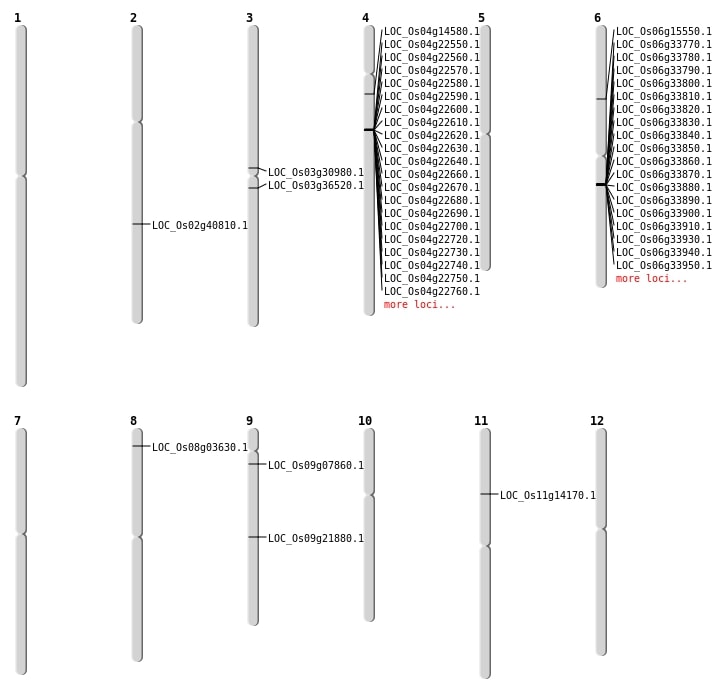
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14137974 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 24292437 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 32088096 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 14019870 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 14886219 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 333003 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 14531398 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 1818193 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 25011806 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 11396642 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 4461271 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 20712721 | T-A | HET | INTRON | |
| SBS | Chr2 | 25996851 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 26172956 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 28087196 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 17645942 | C-T | HET | STOP_GAINED | LOC_Os03g30980 |
| SBS | Chr3 | 30157618 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 7129959 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 7941152 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 12835681 | C-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 24760587 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 4303802 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 971100 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 15072408 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 15419405 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 16290927 | C-T | HET | INTRON | |
| SBS | Chr5 | 1965199 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 20378103 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 14616789 | C-G | HET | UTR_5_PRIME | |
| SBS | Chr6 | 6812272 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 8823064 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g15550 |
| SBS | Chr6 | 9438393 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 11086266 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 12403177 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 15599599 | C-G | HET | INTRON | |
| SBS | Chr7 | 20473417 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 26465630 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 7845546 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 8675827 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 1368032 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 13782991 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 1401781 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 1719664 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g03630 |
| SBS | Chr9 | 13251077 | G-A | HOMO | STOP_GAINED | LOC_Os09g21880 |
| SBS | Chr9 | 3997712 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07860 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 149892 | 149893 | 1 | |
| Deletion | Chr8 | 2305655 | 2305657 | 2 | |
| Deletion | Chr10 | 2763862 | 2763866 | 4 | |
| Deletion | Chr6 | 3212366 | 3212375 | 9 | |
| Deletion | Chr3 | 3444799 | 3444811 | 12 | |
| Deletion | Chr4 | 3526873 | 3526879 | 6 | |
| Deletion | Chr5 | 4680237 | 4680240 | 3 | |
| Deletion | Chr11 | 7914115 | 7914125 | 10 | LOC_Os11g14170 |
| Deletion | Chr4 | 8184028 | 8184033 | 5 | LOC_Os04g14580 |
| Deletion | Chr6 | 8514825 | 8514826 | 1 | |
| Deletion | Chr4 | 12763001 | 12941000 | 178000 | 28 |
| Deletion | Chr10 | 16382612 | 16382648 | 36 | |
| Deletion | Chr11 | 16749360 | 16749407 | 47 | |
| Deletion | Chr6 | 19647001 | 19911000 | 264000 | 39 |
| Deletion | Chr2 | 24733030 | 24733036 | 6 | LOC_Os02g40810 |
| Deletion | Chr7 | 28292358 | 28292359 | 1 | |
| Deletion | Chr4 | 31410576 | 31410579 | 3 | LOC_Os04g52750 |
| Deletion | Chr1 | 31602426 | 31602437 | 11 | |
| Deletion | Chr1 | 40122026 | 40122029 | 3 | |
| Deletion | Chr1 | 40919504 | 40919507 | 3 |
Insertions: 7
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 27644954 | 27959895 | |
| Inversion | Chr6 | 27644979 | 27959908 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 5660128 | Chr3 | 20221656 | LOC_Os03g36520 |
| Translocation | Chr5 | 8562120 | Chr1 | 40306292 |
