Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN873-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN873-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 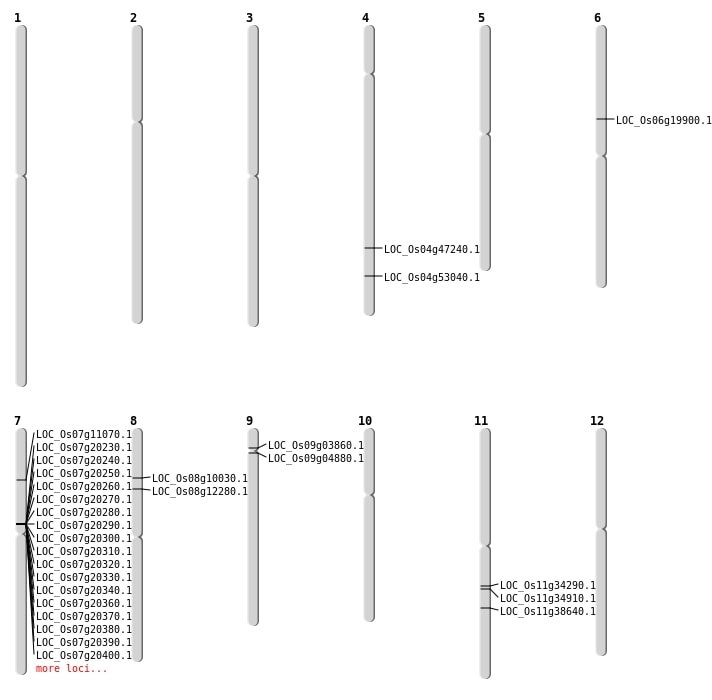
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21118460 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 22419533 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 23332079 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 29394558 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 34489528 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 5276866 | T-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 15468601 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 19626284 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 20130562 | T-G | HOMO | INTRON | |
| SBS | Chr11 | 7029982 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 20898452 | C-T | HET | INTRON | |
| SBS | Chr12 | 22601203 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 13141550 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 2307168 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 25688383 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 26304188 | T-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 21723529 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 495993 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 8413121 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 23624265 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 28053997 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g47240 |
| SBS | Chr4 | 31033662 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 21557849 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 2965101 | A-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 13900523 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g24410 |
| SBS | Chr7 | 15547163 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 19044128 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 19156138 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 22971676 | A-T | HET | INTRON | |
| SBS | Chr7 | 23995263 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 6121432 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g11070 |
| SBS | Chr7 | 8404042 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5812368 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g10030 |
| SBS | Chr8 | 8807498 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 16139103 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 1957343 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 2279495 | C-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1957333 | 1957339 | 6 | LOC_Os09g03860 |
| Deletion | Chr9 | 2600000 | 2600001 | 1 | LOC_Os09g04880 |
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr2 | 4814456 | 4814458 | 2 | |
| Deletion | Chr1 | 5006986 | 5007009 | 23 | |
| Deletion | Chr8 | 7228530 | 7228543 | 13 | LOC_Os08g12280 |
| Deletion | Chr12 | 8142504 | 8142505 | 1 | |
| Deletion | Chr6 | 11382936 | 11382945 | 9 | LOC_Os06g19900 |
| Deletion | Chr7 | 11667001 | 11787000 | 120000 | 18 |
| Deletion | Chr7 | 13347913 | 13347936 | 23 | LOC_Os07g23610 |
| Deletion | Chr7 | 15958468 | 15958469 | 1 | |
| Deletion | Chr7 | 18419110 | 18419112 | 2 | LOC_Os07g31130 |
| Deletion | Chr11 | 18883851 | 18883867 | 16 | |
| Deletion | Chr10 | 19170052 | 19170058 | 6 | |
| Deletion | Chr1 | 20171111 | 20171112 | 1 | |
| Deletion | Chr11 | 20441889 | 20441891 | 2 | |
| Deletion | Chr7 | 21510275 | 21510282 | 7 | LOC_Os07g35950 |
| Deletion | Chr6 | 22242993 | 22243002 | 9 | |
| Deletion | Chr11 | 22910820 | 22910821 | 1 | LOC_Os11g38640 |
| Deletion | Chr7 | 23509062 | 23509063 | 1 | LOC_Os07g39270 |
| Deletion | Chr5 | 25409573 | 25409575 | 2 | |
| Deletion | Chr4 | 25918814 | 25918815 | 1 | |
| Deletion | Chr4 | 25966072 | 25966077 | 5 | |
| Deletion | Chr2 | 35465671 | 35465674 | 3 |
Insertions: 6
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 20044402 | 20092739 | LOC_Os11g34290 |
| Inversion | Chr11 | 20092976 | 20459846 | 2 |
| Inversion | Chr4 | 31095519 | 31597192 | LOC_Os04g53040 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 10902805 | Chr1 | 34373527 | |
| Translocation | Chr11 | 20092962 | Chr10 | 244583 | LOC_Os11g34290 |
