Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN874-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN874-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 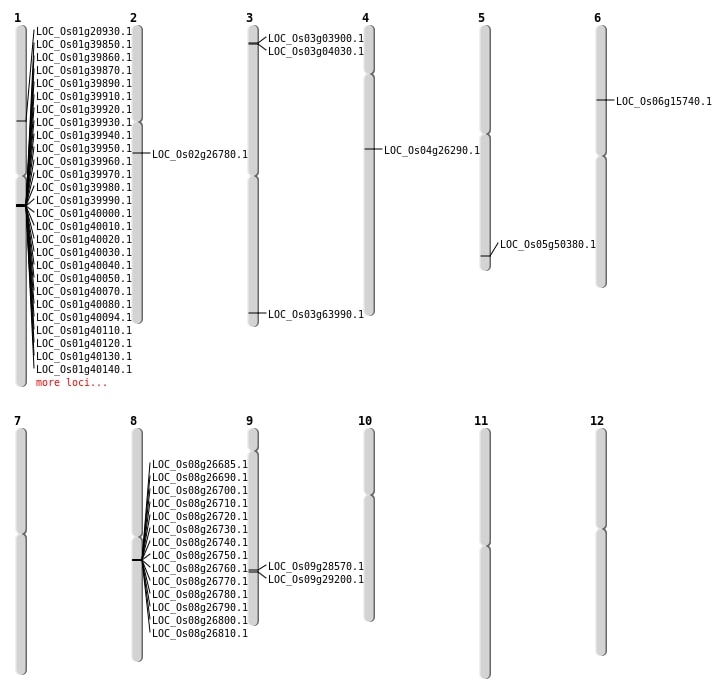
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10442250 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 13207556 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 16621120 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 7958399 | T-G | HET | INTRON | |
| SBS | Chr10 | 8384702 | G-C | HET | INTRON | |
| SBS | Chr11 | 13331718 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 14480752 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 15863252 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 22797420 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 3430711 | T-C | HOMO | INTRON | |
| SBS | Chr12 | 26806536 | C-G | HET | UTR_5_PRIME | |
| SBS | Chr12 | 5564972 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 9096796 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 10199560 | T-A | HET | INTRON | |
| SBS | Chr2 | 11385050 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 20761415 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 21644776 | G-A | HET | INTRON | |
| SBS | Chr2 | 24656822 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 30369138 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 30373141 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 4676840 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 4823894 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 1838424 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g04030 |
| SBS | Chr4 | 15325223 | C-A | HET | STOP_GAINED | LOC_Os04g26290 |
| SBS | Chr4 | 1605980 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 19045630 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 28714245 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 4378758 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 10350035 | A-C | HOMO | INTRON | |
| SBS | Chr5 | 25097864 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 28875115 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g50380 |
| SBS | Chr5 | 67311 | T-A | HET | INTRON | |
| SBS | Chr6 | 25778143 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 6255635 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 6255636 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 14220357 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 16865512 | C-T | HET | INTRON | |
| SBS | Chr7 | 16869100 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr7 | 7101964 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 12530941 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 9227551 | T-C | HOMO | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1776468 | 1776508 | 40 | LOC_Os03g03900 |
| Deletion | Chr3 | 4316789 | 4316802 | 13 | |
| Deletion | Chr9 | 9906706 | 9906707 | 1 | |
| Deletion | Chr7 | 11116628 | 11116637 | 9 | |
| Deletion | Chr8 | 12406568 | 12406571 | 3 | |
| Deletion | Chr9 | 13835705 | 13835714 | 9 | |
| Deletion | Chr6 | 14507639 | 14507649 | 10 | |
| Deletion | Chr3 | 14591687 | 14591702 | 15 | |
| Deletion | Chr6 | 14606073 | 14606090 | 17 | |
| Deletion | Chr7 | 14927863 | 14927873 | 10 | |
| Deletion | Chr8 | 16214001 | 16303000 | 89000 | 14 |
| Deletion | Chr8 | 16240652 | 16261085 | 20433 | 3 |
| Deletion | Chr8 | 16306849 | 16306858 | 9 | |
| Deletion | Chr3 | 19925063 | 19925076 | 13 | |
| Deletion | Chr4 | 21131935 | 21131937 | 2 | |
| Deletion | Chr2 | 21307904 | 21307914 | 10 | |
| Deletion | Chr1 | 22475001 | 22687000 | 212000 | 32 |
| Deletion | Chr8 | 24627845 | 24627850 | 5 | |
| Deletion | Chr4 | 32631509 | 32631514 | 5 | |
| Deletion | Chr1 | 41988472 | 41988603 | 131 | LOC_Os01g72390 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 23388667 | 23388667 | 1 | |
| Insertion | Chr3 | 12521450 | 12521450 | 1 | |
| Insertion | Chr4 | 16564311 | 16564332 | 22 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 11690392 | 22687250 | 2 |
| Inversion | Chr9 | 17382604 | 17747341 | 2 |
| Inversion | Chr3 | 35283293 | 36169206 | LOC_Os03g63990 |
| Inversion | Chr3 | 35283304 | 35324960 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2691428 | Chr6 | 14189667 | |
| Translocation | Chr2 | 15723259 | Chr1 | 9625513 | LOC_Os02g26780 |
| Translocation | Chr11 | 27773717 | Chr6 | 8926366 | LOC_Os06g15740 |
