Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN883-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN883-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 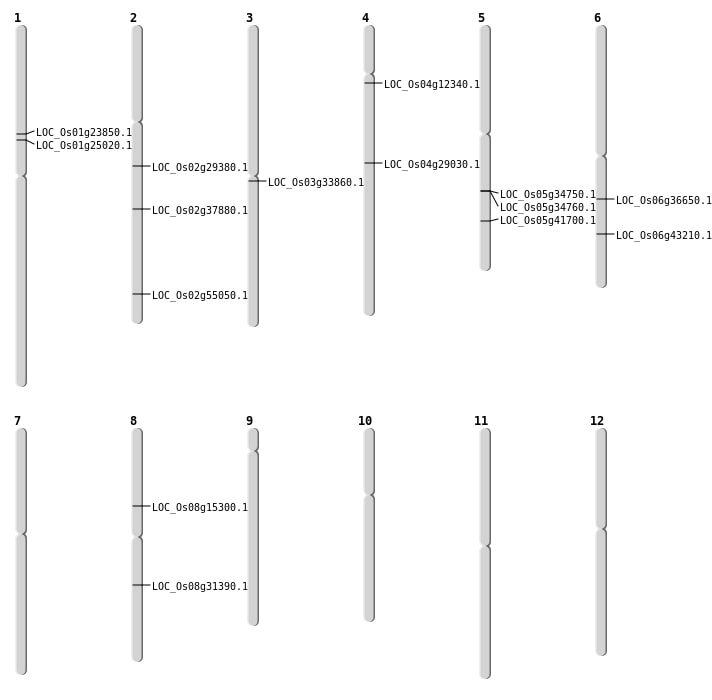
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14347753 | G-A | HOMO | INTRON | |
| SBS | Chr1 | 29910189 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 30327032 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 3550238 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 3550239 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 41612971 | C-T | HET | INTRON | |
| SBS | Chr1 | 9632093 | C-G | HOMO | INTRON | |
| SBS | Chr10 | 3204817 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 15323127 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 28034651 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 31627856 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 33722931 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g55050 |
| SBS | Chr2 | 33743162 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 4276599 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 33876436 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 6729210 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 14963203 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 16745064 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21538333 | G-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 22513683 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 2314837 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 23179747 | T-G | HET | INTRON | |
| SBS | Chr4 | 27607057 | G-A | HET | INTRON | |
| SBS | Chr4 | 7120580 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 14340270 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 21538960 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g36650 |
| SBS | Chr6 | 26780417 | G-T | HET | INTRON | |
| SBS | Chr6 | 5614859 | A-T | HET | INTRON | |
| SBS | Chr6 | 8584180 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 13304769 | C-T | HET | INTRON | |
| SBS | Chr7 | 23893021 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 12652696 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 15429262 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 15429263 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 16057415 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 19406897 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g31390 |
| SBS | Chr8 | 9434679 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 13433707 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 21775163 | A-T | HOMO | INTRON | |
| SBS | Chr9 | 21997963 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5738713 | A-C | HOMO | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 435534 | 435607 | 73 | |
| Deletion | Chr12 | 1263205 | 1263216 | 11 | |
| Deletion | Chr12 | 1823471 | 1823476 | 5 | |
| Deletion | Chr8 | 4198399 | 4198551 | 152 | |
| Deletion | Chr1 | 4799985 | 4799991 | 6 | |
| Deletion | Chr1 | 5325906 | 5325912 | 6 | |
| Deletion | Chr9 | 5533556 | 5533559 | 3 | |
| Deletion | Chr4 | 6809054 | 6809813 | 759 | LOC_Os04g12340 |
| Deletion | Chr2 | 7531569 | 7531570 | 1 | |
| Deletion | Chr6 | 8739278 | 8739289 | 11 | |
| Deletion | Chr1 | 13423297 | 13423300 | 3 | LOC_Os01g23850 |
| Deletion | Chr9 | 15661634 | 15661643 | 9 | |
| Deletion | Chr4 | 17201340 | 17201347 | 7 | LOC_Os04g29030 |
| Deletion | Chr7 | 19429672 | 19429687 | 15 | |
| Deletion | Chr12 | 20349003 | 20349006 | 3 | |
| Deletion | Chr5 | 20611001 | 20622000 | 11000 | 2 |
| Deletion | Chr2 | 22893690 | 22893695 | 5 | LOC_Os02g37880 |
| Deletion | Chr5 | 23535834 | 23535839 | 5 | |
| Deletion | Chr6 | 23953122 | 23953130 | 8 | |
| Deletion | Chr4 | 25051234 | 25051236 | 2 | |
| Deletion | Chr6 | 25968065 | 25968069 | 4 | LOC_Os06g43210 |
| Deletion | Chr1 | 29490289 | 29490296 | 7 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 14113580 | 14113580 | 1 | LOC_Os01g25020 |
| Insertion | Chr1 | 19331298 | 19331299 | 2 | |
| Insertion | Chr10 | 13066275 | 13066275 | 1 | |
| Insertion | Chr2 | 17454917 | 17454918 | 2 | LOC_Os02g29380 |
| Insertion | Chr5 | 14656419 | 14656419 | 1 | |
| Insertion | Chr5 | 20428418 | 20428418 | 1 | |
| Insertion | Chr9 | 8191471 | 8191472 | 2 |
No Inversion
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 5065839 | Chr5 | 24396057 | LOC_Os05g41700 |
| Translocation | Chr9 | 5066412 | Chr8 | 26485687 | |
| Translocation | Chr7 | 6399806 | Chr5 | 10232217 | |
| Translocation | Chr8 | 9306273 | Chr3 | 19340951 | 2 |
| Translocation | Chr7 | 15285313 | Chr2 | 15956328 | |
| Translocation | Chr3 | 32005350 | Chr1 | 42745584 |
