Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN886-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN886-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 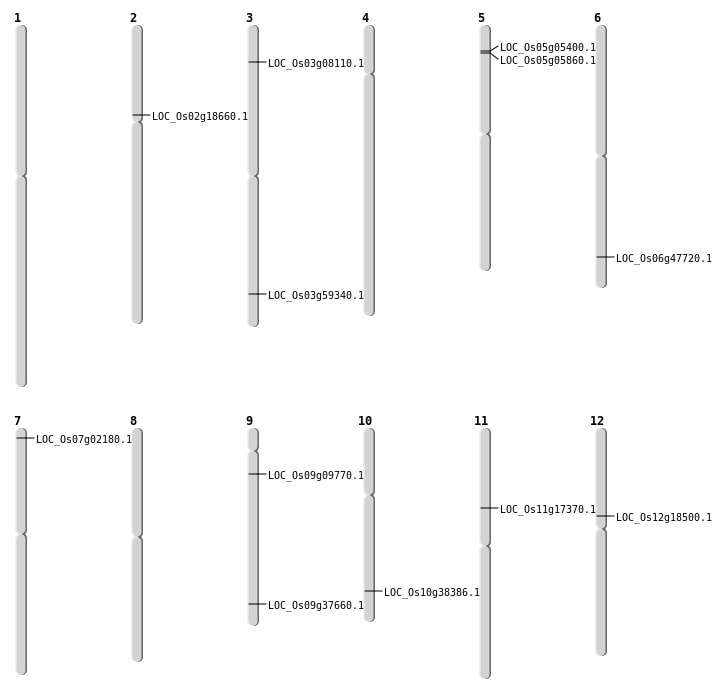
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13414082 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 26767897 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr1 | 27976326 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 20512195 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g38386 |
| SBS | Chr11 | 15706879 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 22999473 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 23387748 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 9680728 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g17370 |
| SBS | Chr12 | 19541031 | T-C | HET | INTRON | |
| SBS | Chr12 | 23593232 | G-A | HET | INTRON | |
| SBS | Chr2 | 10399803 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 14148541 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 17533721 | C-G | HET | INTRON | |
| SBS | Chr2 | 17865253 | A-G | HET | INTRON | |
| SBS | Chr2 | 23691737 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 27043449 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 10429488 | C-T | HET | INTRON | |
| SBS | Chr3 | 14101827 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 15756800 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 28490869 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 33784090 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g59340 |
| SBS | Chr4 | 11609536 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 33348689 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 8912519 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 16929493 | G-A | HET | INTRON | |
| SBS | Chr5 | 28797131 | T-A | HET | INTRON | |
| SBS | Chr6 | 22973955 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 24280416 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 28976808 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 12456921 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 12456922 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 28553160 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 693725 | A-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g02180 |
| SBS | Chr7 | 9154182 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 15423636 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 28217997 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 19209660 | G-A | HOMO | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2271564 | 2271566 | 2 | |
| Deletion | Chr5 | 2684297 | 2684305 | 8 | LOC_Os05g05400 |
| Deletion | Chr5 | 2939875 | 2940701 | 826 | LOC_Os05g05860 |
| Deletion | Chr1 | 3547454 | 3547456 | 2 | |
| Deletion | Chr2 | 3999743 | 3999750 | 7 | |
| Deletion | Chr4 | 4291204 | 4291221 | 17 | |
| Deletion | Chr5 | 4633018 | 4633023 | 5 | |
| Deletion | Chr9 | 5294523 | 5294608 | 85 | LOC_Os09g09770 |
| Deletion | Chr8 | 8350191 | 8350194 | 3 | |
| Deletion | Chr12 | 10696206 | 10696213 | 7 | LOC_Os12g18500 |
| Deletion | Chr6 | 15540733 | 15540739 | 6 | |
| Deletion | Chr6 | 15707867 | 15707869 | 2 | |
| Deletion | Chr2 | 16288489 | 16288496 | 7 | |
| Deletion | Chr5 | 16930761 | 16930771 | 10 | |
| Deletion | Chr11 | 18736587 | 18736588 | 1 | |
| Deletion | Chr10 | 20509471 | 20509472 | 1 | |
| Deletion | Chr9 | 21703884 | 21703889 | 5 | LOC_Os09g37660 |
| Deletion | Chr7 | 22982172 | 22982180 | 8 | |
| Deletion | Chr3 | 24403602 | 24403614 | 12 | |
| Deletion | Chr1 | 25611857 | 25611863 | 6 | |
| Deletion | Chr6 | 25766305 | 25766306 | 1 | |
| Deletion | Chr8 | 28353987 | 28354011 | 24 | |
| Deletion | Chr8 | 28373668 | 28373672 | 4 | |
| Deletion | Chr6 | 28538125 | 28538136 | 11 | |
| Deletion | Chr11 | 28657916 | 28657917 | 1 | |
| Deletion | Chr6 | 28876681 | 28876697 | 16 | LOC_Os06g47720 |
| Deletion | Chr7 | 29529257 | 29529259 | 2 | |
| Deletion | Chr2 | 31436769 | 31436781 | 12 | |
| Deletion | Chr1 | 33460154 | 33460163 | 9 | |
| Deletion | Chr1 | 40694301 | 40694302 | 1 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 25828609 | 25828616 | 8 | |
| Insertion | Chr6 | 7855242 | 7855243 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 8926190 | 10872630 | LOC_Os02g18660 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 4141742 | Chr2 | 17320663 | LOC_Os03g08110 |
