Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN889-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN889-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 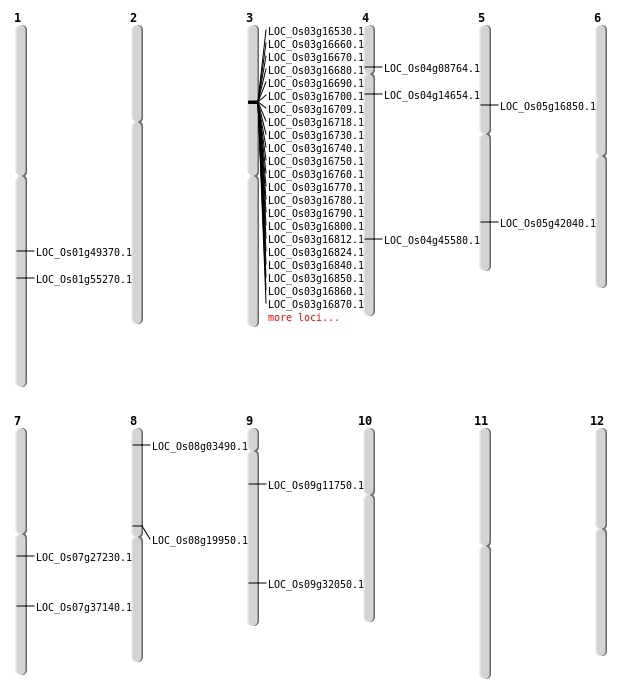
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13019815 | A-G | HET | INTRON | |
| SBS | Chr1 | 22203196 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 28948701 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 31811657 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g55270 |
| SBS | Chr10 | 15824395 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 27712752 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 21925443 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 10301591 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 20842851 | C-A | HOMO | INTRON | |
| SBS | Chr3 | 17369719 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 23061079 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 9112407 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g16530 |
| SBS | Chr4 | 13909426 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 17196123 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 17676636 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 32714081 | C-G | HET | INTRON | |
| SBS | Chr4 | 6136966 | T-G | HET | INTRON | |
| SBS | Chr5 | 24466092 | C-G | HET | INTRON | |
| SBS | Chr6 | 14268783 | G-A | HET | INTRON | |
| SBS | Chr6 | 18113562 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 14341554 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 14341555 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 15821097 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g27230 |
| SBS | Chr7 | 15821098 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 18925970 | T-C | HET | INTRON | |
| SBS | Chr7 | 22248973 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g37140 |
| SBS | Chr7 | 2953033 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 11942052 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g19950 |
| SBS | Chr8 | 1634938 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g03490 |
| SBS | Chr8 | 3961080 | A-T | HET | INTRON | |
| SBS | Chr9 | 21185709 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 3186030 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 6569849 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g11750 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 460307 | 460308 | 1 | |
| Deletion | Chr12 | 4684832 | 4684838 | 6 | |
| Deletion | Chr4 | 4770179 | 4770193 | 14 | LOC_Os04g08764 |
| Deletion | Chr11 | 4792616 | 4792635 | 19 | |
| Deletion | Chr5 | 7705220 | 7705228 | 8 | |
| Deletion | Chr11 | 7716129 | 7716137 | 8 | |
| Deletion | Chr4 | 8216847 | 8216851 | 4 | LOC_Os04g14654 |
| Deletion | Chr3 | 9197001 | 9417000 | 220000 | 27 |
| Deletion | Chr5 | 9606094 | 9606101 | 7 | LOC_Os05g16850 |
| Deletion | Chr9 | 12143484 | 12143485 | 1 | |
| Deletion | Chr4 | 13119784 | 13119795 | 11 | |
| Deletion | Chr6 | 14268645 | 14268676 | 31 | |
| Deletion | Chr8 | 16048967 | 16048968 | 1 | |
| Deletion | Chr9 | 19126512 | 19126513 | 1 | LOC_Os09g32050 |
| Deletion | Chr2 | 19909968 | 19909970 | 2 | |
| Deletion | Chr11 | 24269257 | 24269272 | 15 | |
| Deletion | Chr5 | 24607046 | 24607071 | 25 | LOC_Os05g42040 |
| Deletion | Chr8 | 26647492 | 26647496 | 4 | |
| Deletion | Chr1 | 28381403 | 28381414 | 11 | LOC_Os01g49370 |
| Deletion | Chr11 | 28951144 | 28951152 | 8 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 26967544 | 31371732 | LOC_Os04g45580 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 23038834 | Chr3 | 20268724 | LOC_Os03g36560 |
