Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN899-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN899-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 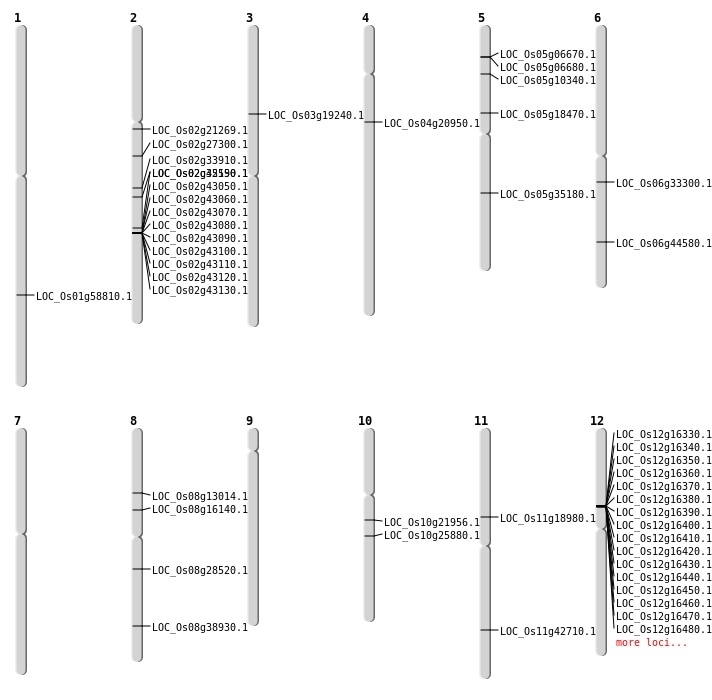
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13467976 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 31394305 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 33655492 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11784218 | G-C | HET | INTERGENIC | |
| SBS | Chr10 | 11784219 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 12407415 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 17292057 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 11179822 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 1167693 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 18258937 | T-G | HET | INTRON | |
| SBS | Chr11 | 25728381 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g42710 |
| SBS | Chr11 | 7589924 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 10496081 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g18190 |
| SBS | Chr12 | 21403139 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 11685367 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 14509345 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr2 | 20553194 | C-A | HET | INTRON | |
| SBS | Chr2 | 20553195 | T-A | HET | INTRON | |
| SBS | Chr2 | 26392004 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 12212667 | A-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 21448232 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 36208168 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 5435212 | A-G | HET | INTRON | |
| SBS | Chr4 | 10793252 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 21910966 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 22977879 | T-C | HOMO | UTR_5_PRIME | |
| SBS | Chr4 | 6168483 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20891404 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g35180 |
| SBS | Chr5 | 21516761 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 6784789 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 13623073 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 18226425 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 17418744 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g28520 |
| SBS | Chr8 | 24603088 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g38930 |
| SBS | Chr8 | 4854231 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 4854232 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 10926085 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 2774317 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 4897193 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 9612539 | C-T | HET | INTERGENIC |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 2881901 | 2881918 | 17 | |
| Deletion | Chr12 | 3389438 | 3389439 | 1 | |
| Deletion | Chr7 | 4537088 | 4537090 | 2 | |
| Deletion | Chr7 | 4551120 | 4551122 | 2 | |
| Deletion | Chr5 | 6784790 | 6784792 | 2 | |
| Deletion | Chr5 | 6784792 | 6784794 | 2 | |
| Deletion | Chr12 | 9341001 | 9485000 | 144000 | 24 |
| Deletion | Chr9 | 10133934 | 10133939 | 5 | |
| Deletion | Chr3 | 11543836 | 11543844 | 8 | |
| Deletion | Chr10 | 11785830 | 11785832 | 2 | |
| Deletion | Chr5 | 12348308 | 12348320 | 12 | |
| Deletion | Chr3 | 12370005 | 12370006 | 1 | |
| Deletion | Chr9 | 13311171 | 13311173 | 2 | |
| Deletion | Chr10 | 13415588 | 13415589 | 1 | LOC_Os10g25880 |
| Deletion | Chr4 | 13779622 | 13779623 | 1 | |
| Deletion | Chr4 | 16117707 | 16117715 | 8 | |
| Deletion | Chr9 | 16300272 | 16300273 | 1 | |
| Deletion | Chr11 | 17383154 | 17383167 | 13 | |
| Deletion | Chr6 | 17734589 | 17734591 | 2 | |
| Deletion | Chr6 | 19388027 | 19388040 | 13 | LOC_Os06g33300 |
| Deletion | Chr2 | 20207848 | 20207869 | 21 | LOC_Os02g33910 |
| Deletion | Chr2 | 21411014 | 21411037 | 23 | LOC_Os02g35590 |
| Deletion | Chr11 | 23872173 | 23872174 | 1 | |
| Deletion | Chr2 | 25922001 | 25976000 | 54000 | 9 |
| Deletion | Chr5 | 27922668 | 27922676 | 8 | |
| Deletion | Chr7 | 28372205 | 28372214 | 9 | |
| Deletion | Chr1 | 30258509 | 30258510 | 1 | LOC_Os01g52640 |
| Deletion | Chr1 | 30576706 | 30576708 | 2 | |
| Deletion | Chr1 | 31394307 | 31394311 | 4 | |
| Deletion | Chr4 | 33061599 | 33061604 | 5 | |
| Deletion | Chr1 | 33982501 | 33982504 | 3 | LOC_Os01g58810 |
| Deletion | Chr2 | 35438290 | 35438294 | 4 | |
| Deletion | Chr1 | 39069905 | 39069911 | 6 |
Insertions: 5
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 3448868 | 4208402 | |
| Inversion | Chr5 | 3465852 | 4208399 | LOC_Os05g06670 |
| Inversion | Chr4 | 6100417 | 18323903 | |
| Inversion | Chr2 | 16081489 | 16096151 | |
| Inversion | Chr2 | 16082615 | 16096152 | LOC_Os02g27300 |
| Inversion | Chr10 | 20129261 | 20209851 | |
| Inversion | Chr2 | 25348487 | 28249280 | LOC_Os02g42150 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 5744942 | Chr2 | 16081486 | |
| Translocation | Chr8 | 7731011 | Chr2 | 12621038 | 2 |
| Translocation | Chr11 | 8958664 | Chr5 | 10674037 | LOC_Os05g18470 |
| Translocation | Chr8 | 9841284 | Chr3 | 18929725 | LOC_Os08g16140 |
| Translocation | Chr11 | 10816576 | Chr3 | 10819053 | 2 |
| Translocation | Chr10 | 11338878 | Chr5 | 5644610 | 2 |
| Translocation | Chr7 | 20719776 | Chr5 | 3465867 | LOC_Os05g06670 |
| Translocation | Chr7 | 20720233 | Chr5 | 3468016 | LOC_Os05g06680 |
| Translocation | Chr6 | 26920594 | Chr4 | 11757204 | 2 |
| Translocation | Chr6 | 29009429 | Chr2 | 16082677 | LOC_Os02g27300 |
