Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN900-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN900-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 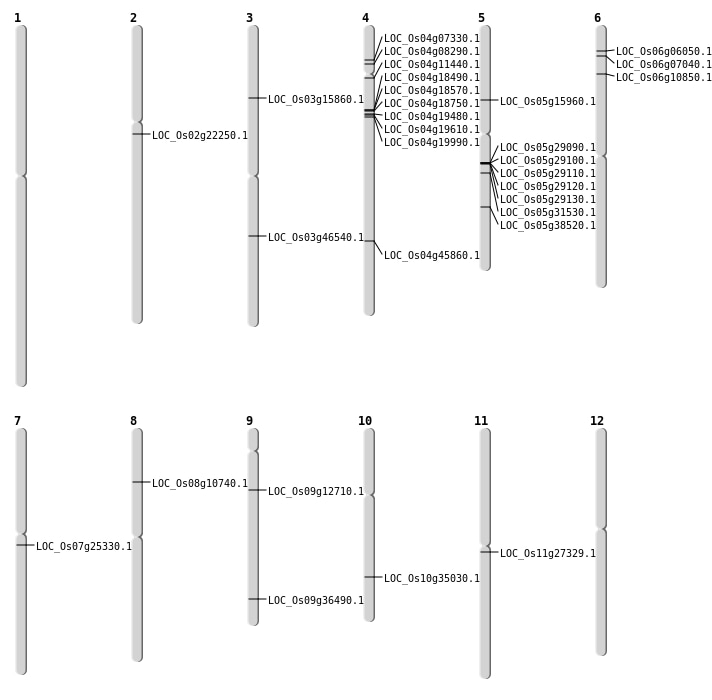
|
| Variant Information |
|---|
Single base substitutions: 21
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 529345 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 19022673 | T-A | HET | INTRON | |
| SBS | Chr10 | 19026145 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 22905633 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 3056656 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 7593078 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 35185487 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 10847702 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g19480 |
| SBS | Chr4 | 12850652 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 13486002 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 4423778 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 4423781 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 9917214 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 22774621 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 10467810 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 14467536 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g25330 |
| SBS | Chr7 | 22710155 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5412885 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 6321438 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g10740 |
| SBS | Chr9 | 3559707 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 8258080 | G-A | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1754144 | 1754153 | 9 | |
| Deletion | Chr5 | 1763714 | 1763715 | 1 | |
| Deletion | Chr8 | 3565913 | 3565924 | 11 | |
| Deletion | Chr3 | 4316260 | 4316262 | 2 | |
| Deletion | Chr4 | 4346644 | 4346738 | 94 | |
| Deletion | Chr6 | 5658686 | 5658999 | 313 | |
| Deletion | Chr4 | 6257287 | 6257888 | 601 | LOC_Os04g11440 |
| Deletion | Chr9 | 7271303 | 7271304 | 1 | LOC_Os09g12710 |
| Deletion | Chr4 | 9933087 | 9933096 | 9 | |
| Deletion | Chr3 | 10288965 | 10288966 | 1 | |
| Deletion | Chr4 | 10404054 | 10404065 | 11 | LOC_Os04g18750 |
| Deletion | Chr1 | 11966025 | 11966033 | 8 | |
| Deletion | Chr2 | 13267214 | 13267217 | 3 | LOC_Os02g22250 |
| Deletion | Chr5 | 17082001 | 17118000 | 36000 | 5 |
| Deletion | Chr5 | 18346421 | 18349627 | 3206 | LOC_Os05g31530 |
| Deletion | Chr10 | 18692324 | 18692333 | 9 | LOC_Os10g35030 |
| Deletion | Chr9 | 21055737 | 21055742 | 5 | LOC_Os09g36490 |
| Deletion | Chr1 | 22063574 | 22063581 | 7 | |
| Deletion | Chr3 | 23295532 | 23295536 | 4 | |
| Deletion | Chr1 | 24652833 | 24652835 | 2 | |
| Deletion | Chr8 | 26113193 | 26113198 | 5 | |
| Deletion | Chr3 | 26332356 | 26336912 | 4556 | LOC_Os03g46540 |
| Deletion | Chr4 | 27157611 | 27157747 | 136 | LOC_Os04g45860 |
Insertions: 7
Inversions: 10
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 2782177 | 3345947 | 2 |
| Inversion | Chr6 | 2782217 | 3345947 | 2 |
| Inversion | Chr4 | 3900278 | 4425123 | 2 |
| Inversion | Chr4 | 10225299 | 10765458 | LOC_Os04g18490 |
| Inversion | Chr4 | 10253305 | 10767864 | LOC_Os04g18570 |
| Inversion | Chr4 | 10765455 | 10924108 | LOC_Os04g19610 |
| Inversion | Chr4 | 10823394 | 11145041 | LOC_Os04g19990 |
| Inversion | Chr4 | 10824712 | 11145453 | LOC_Os04g19990 |
| Inversion | Chr5 | 17082221 | 18349624 | LOC_Os05g29090 |
| Inversion | Chr5 | 17118247 | 18346447 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 5658983 | Chr3 | 8751633 | 2 |
| Translocation | Chr11 | 11359049 | Chr5 | 9013241 | LOC_Os05g15960 |
| Translocation | Chr11 | 15714467 | Chr9 | 6778101 | LOC_Os11g27329 |
| Translocation | Chr5 | 17118222 | Chr4 | 6257316 | 2 |
| Translocation | Chr11 | 17512065 | Chr7 | 13570804 | |
| Translocation | Chr5 | 22598187 | Chr4 | 6257934 | 2 |
| Translocation | Chr8 | 22804306 | Chr2 | 16275328 |
