Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN914-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN914-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 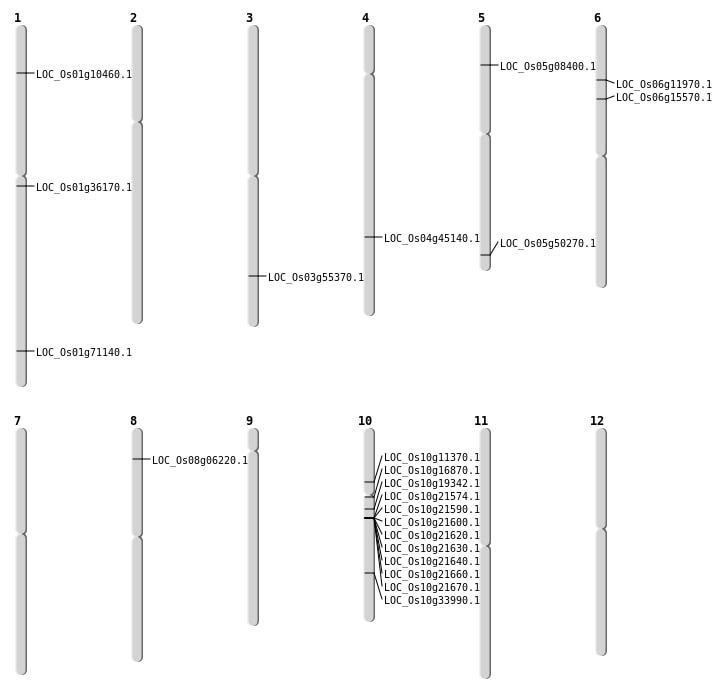
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10409185 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 26045957 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 26762137 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 26909736 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 30609462 | A-G | HET | INTRON | |
| SBS | Chr1 | 37869092 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 37869093 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 16570207 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 17400609 | T-C | HET | INTRON | |
| SBS | Chr10 | 6304367 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g11370 |
| SBS | Chr10 | 6526349 | A-T | HET | INTRON | |
| SBS | Chr11 | 4314135 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 9540247 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 23353027 | G-T | HET | INTRON | |
| SBS | Chr12 | 8706389 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 8706390 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 14866176 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 26164165 | T-C | HOMO | UTR_5_PRIME | |
| SBS | Chr3 | 16831602 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 18290959 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 23491897 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 23552634 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 31501564 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g55370 |
| SBS | Chr4 | 1724403 | T-C | HET | INTRON | |
| SBS | Chr5 | 24724013 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 26386722 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr5 | 29714978 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 4582177 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g08400 |
| SBS | Chr6 | 8366028 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 10575173 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 9300138 | C-T | HOMO | INTRON | |
| SBS | Chr9 | 10798085 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 20008035 | C-A | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 5272579 | 5272583 | 4 | |
| Deletion | Chr1 | 5530582 | 5530587 | 5 | LOC_Os01g10460 |
| Deletion | Chr6 | 6372529 | 6372532 | 3 | LOC_Os06g11970 |
| Deletion | Chr10 | 8408305 | 8408310 | 5 | LOC_Os10g16870 |
| Deletion | Chr6 | 8829542 | 8829551 | 9 | LOC_Os06g15570 |
| Deletion | Chr4 | 8902052 | 8902056 | 4 | |
| Deletion | Chr10 | 10695472 | 10695488 | 16 | |
| Deletion | Chr10 | 11049001 | 11115000 | 66000 | 8 |
| Deletion | Chr6 | 12029020 | 12029043 | 23 | |
| Deletion | Chr9 | 12630321 | 12630325 | 4 | |
| Deletion | Chr11 | 13948589 | 13948600 | 11 | |
| Deletion | Chr6 | 14158631 | 14158643 | 12 | |
| Deletion | Chr6 | 14200943 | 14200944 | 1 | |
| Deletion | Chr10 | 15299893 | 15300044 | 151 | |
| Deletion | Chr2 | 15595112 | 15595115 | 3 | |
| Deletion | Chr10 | 17310374 | 17310376 | 2 | |
| Deletion | Chr10 | 18129752 | 18129758 | 6 | LOC_Os10g33990 |
| Deletion | Chr5 | 18762253 | 18762274 | 21 | |
| Deletion | Chr3 | 23625378 | 23625400 | 22 | |
| Deletion | Chr8 | 24209315 | 24209336 | 21 | |
| Deletion | Chr4 | 26691234 | 26691237 | 3 | LOC_Os04g45140 |
| Deletion | Chr6 | 27684273 | 27684283 | 10 | |
| Deletion | Chr1 | 28394213 | 28394219 | 6 | |
| Deletion | Chr5 | 28817833 | 28817844 | 11 | LOC_Os05g50270 |
| Deletion | Chr4 | 30867905 | 30867906 | 1 | |
| Deletion | Chr3 | 31522308 | 31522309 | 1 | |
| Deletion | Chr3 | 31854791 | 31854794 | 3 | |
| Deletion | Chr1 | 33714992 | 33714997 | 5 | |
| Deletion | Chr4 | 33744511 | 33744519 | 8 | |
| Deletion | Chr1 | 41170450 | 41170454 | 4 | LOC_Os01g71140 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 20006690 | 20006691 | 2 | LOC_Os01g36170 |
| Insertion | Chr2 | 19578310 | 19578310 | 1 | |
| Insertion | Chr5 | 19049178 | 19049179 | 2 | |
| Insertion | Chr7 | 13242113 | 13242114 | 2 | |
| Insertion | Chr8 | 20131861 | 20131862 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 3367392 | 3431959 | 2 |
| Inversion | Chr12 | 7651724 | 7901982 | |
| Inversion | Chr10 | 8869142 | 11049274 | |
| Inversion | Chr10 | 9880192 | 11132768 | LOC_Os10g19342 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 7651761 | Chr10 | 15300112 | |
| Translocation | Chr12 | 7901981 | Chr10 | 15299894 | |
| Translocation | Chr3 | 32459922 | Chr1 | 26315693 |
