Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN919-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN919-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 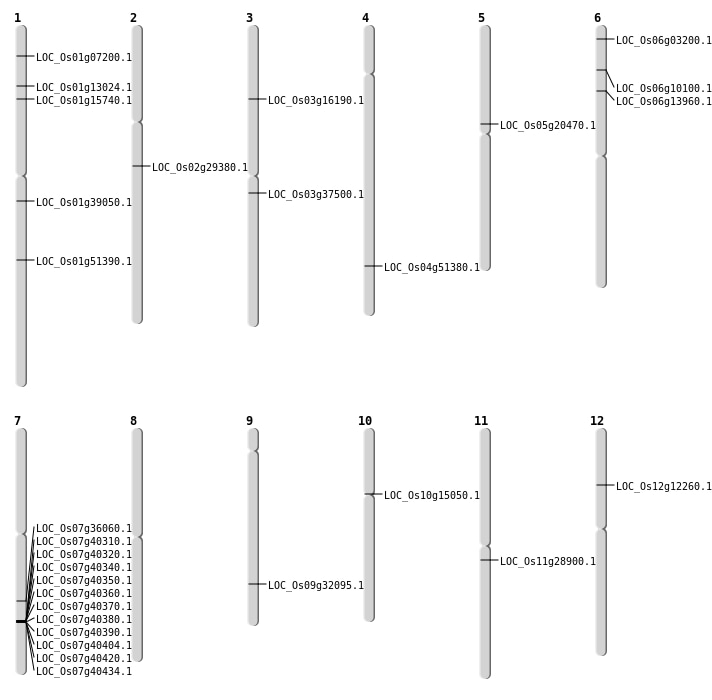
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21264982 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 24830361 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 32793329 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 33798088 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 35731286 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 36065994 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 11386778 | G-A | HET | INTRON | |
| SBS | Chr10 | 11386779 | C-A | HET | INTRON | |
| SBS | Chr10 | 12300840 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 482984 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 14914276 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8896185 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 398810 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 8640412 | A-C | HET | INTRON | |
| SBS | Chr2 | 26170850 | G-A | HET | INTRON | |
| SBS | Chr2 | 26935420 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 27920633 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 18548193 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 19239660 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 19329637 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 28530674 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 8927780 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g16190 |
| SBS | Chr4 | 15882627 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 30429484 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g51380 |
| SBS | Chr4 | 8188421 | T-C | HET | INTRON | |
| SBS | Chr5 | 13678420 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 29262486 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 10946066 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 13451574 | C-T | HET | INTRON | |
| SBS | Chr6 | 21359061 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 25472902 | G-A | HET | INTRON | |
| SBS | Chr6 | 8704932 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 9644612 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 16599335 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 20629402 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 21565052 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36060 |
| SBS | Chr7 | 24005781 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 4421633 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 5965408 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 3072523 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 8894933 | C-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 19158508 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 19158509 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g32095 |
| SBS | Chr9 | 20980101 | T-C | HET | INTRON |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 159515 | 159526 | 11 | |
| Deletion | Chr8 | 2927123 | 2927126 | 3 | |
| Deletion | Chr1 | 3397528 | 3397529 | 1 | LOC_Os01g07200 |
| Deletion | Chr7 | 4490445 | 4490447 | 2 | |
| Deletion | Chr6 | 5143758 | 5143768 | 10 | LOC_Os06g10100 |
| Deletion | Chr6 | 6196247 | 6196251 | 4 | |
| Deletion | Chr12 | 6730973 | 6730977 | 4 | LOC_Os12g12260 |
| Deletion | Chr5 | 7957321 | 7957322 | 1 | |
| Deletion | Chr5 | 8629753 | 8629754 | 1 | |
| Deletion | Chr12 | 9286229 | 9286230 | 1 | |
| Deletion | Chr11 | 13493323 | 13493334 | 11 | |
| Deletion | Chr5 | 13738221 | 13738258 | 37 | |
| Deletion | Chr2 | 15599807 | 15599827 | 20 | |
| Deletion | Chr3 | 16410504 | 16410513 | 9 | |
| Deletion | Chr2 | 17450369 | 17450843 | 474 | LOC_Os02g29380 |
| Deletion | Chr5 | 17800891 | 17800915 | 24 | |
| Deletion | Chr11 | 19977005 | 19977006 | 1 | |
| Deletion | Chr3 | 20812322 | 20812331 | 9 | LOC_Os03g37500 |
| Deletion | Chr1 | 21954699 | 21954715 | 16 | LOC_Os01g39050 |
| Deletion | Chr9 | 22655673 | 22655680 | 7 | |
| Deletion | Chr3 | 22958045 | 22958051 | 6 | |
| Deletion | Chr3 | 23080036 | 23080068 | 32 | |
| Deletion | Chr3 | 23332547 | 23332552 | 5 | |
| Deletion | Chr2 | 23874085 | 23874088 | 3 | |
| Deletion | Chr7 | 24173001 | 24237000 | 64000 | 11 |
| Deletion | Chr7 | 25701209 | 25701216 | 7 | |
| Deletion | Chr5 | 26797561 | 26797563 | 2 | |
| Deletion | Chr5 | 27231318 | 27231319 | 1 | |
| Deletion | Chr7 | 27422944 | 27422954 | 10 | |
| Deletion | Chr5 | 28233941 | 28233948 | 7 | |
| Deletion | Chr4 | 28925767 | 28925771 | 4 | |
| Deletion | Chr1 | 29546193 | 29546209 | 16 | LOC_Os01g51390 |
| Deletion | Chr3 | 34116357 | 34116363 | 6 |
Insertions: 10
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 6640205 | 6719172 | |
| Inversion | Chr12 | 6640221 | 6719185 | |
| Inversion | Chr1 | 7245364 | 8854939 | 2 |
| Inversion | Chr1 | 7286531 | 8854935 | LOC_Os01g15740 |
| Inversion | Chr1 | 10060500 | 10906066 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 1208008 | Chr5 | 12013402 | 2 |
| Translocation | Chr6 | 11983751 | Chr4 | 12439019 | |
| Translocation | Chr3 | 13030451 | Chr1 | 7297269 | |
| Translocation | Chr3 | 13030460 | Chr1 | 7287550 | |
| Translocation | Chr11 | 15308530 | Chr6 | 30259199 | |
| Translocation | Chr11 | 16725353 | Chr10 | 7881784 | 2 |
| Translocation | Chr7 | 24172638 | Chr1 | 7447676 | LOC_Os07g40310 |
| Translocation | Chr7 | 24237291 | Chr1 | 7470465 | LOC_Os07g40434 |
| Translocation | Chr11 | 24831650 | Chr6 | 7782063 | LOC_Os06g13960 |
