Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN939-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN939-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 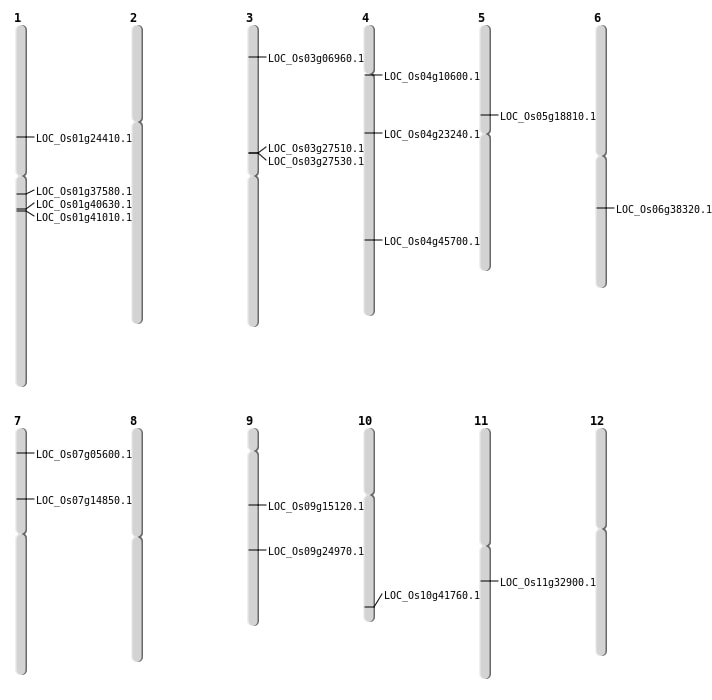
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13762378 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g24410 |
| SBS | Chr1 | 14244358 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 19862288 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 25913578 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 42314248 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 6363796 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 10426270 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 15968712 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 22476491 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g41760 |
| SBS | Chr10 | 672882 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 17287190 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 17287191 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 24270495 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 24273337 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 26547676 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 25815612 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr2 | 28736446 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 4592200 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 12071705 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 12705674 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 26938960 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 29329600 | C-T | HET | INTRON | |
| SBS | Chr3 | 30769790 | G-T | HET | INTRON | |
| SBS | Chr3 | 30769792 | C-A | HET | INTRON | |
| SBS | Chr3 | 30769794 | A-T | HET | INTRON | |
| SBS | Chr3 | 4347645 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 13262006 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g23240 |
| SBS | Chr4 | 5757085 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g10600 |
| SBS | Chr5 | 16897098 | T-A | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 17840492 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 3822719 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 14970087 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 26658106 | T-A | HET | INTRON | |
| SBS | Chr7 | 10808021 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 20500167 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 2630111 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g05600 |
| SBS | Chr7 | 4395036 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 1351532 | A-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 13667458 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 13667464 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 367106 | T-A | HET | INTERGENIC |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 3987115 | 3987116 | 1 | |
| Deletion | Chr12 | 7793895 | 7793896 | 1 | |
| Deletion | Chr9 | 9178013 | 9178024 | 11 | LOC_Os09g15120 |
| Deletion | Chr5 | 10931697 | 10931698 | 1 | LOC_Os05g18810 |
| Deletion | Chr1 | 12699874 | 12699879 | 5 | |
| Deletion | Chr9 | 14918266 | 14918272 | 6 | LOC_Os09g24970 |
| Deletion | Chr7 | 15344151 | 15344160 | 9 | |
| Deletion | Chr5 | 16598855 | 16598874 | 19 | |
| Deletion | Chr9 | 18130730 | 18130961 | 231 | |
| Deletion | Chr7 | 18846637 | 18846638 | 1 | |
| Deletion | Chr5 | 19424100 | 19424101 | 1 | |
| Deletion | Chr11 | 19437951 | 19437953 | 2 | LOC_Os11g32900 |
| Deletion | Chr4 | 19682462 | 19682465 | 3 | |
| Deletion | Chr1 | 21013225 | 21013353 | 128 | LOC_Os01g37580 |
| Deletion | Chr6 | 25029418 | 25029428 | 10 | |
| Deletion | Chr11 | 25387235 | 25387240 | 5 | |
| Deletion | Chr11 | 25525973 | 25525974 | 1 | |
| Deletion | Chr4 | 27034352 | 27034353 | 1 | LOC_Os04g45700 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 27711045 | 27711050 | 6 | |
| Insertion | Chr2 | 21771740 | 21771741 | 2 | |
| Insertion | Chr3 | 5260532 | 5260535 | 4 | |
| Insertion | Chr7 | 8499708 | 8499710 | 3 | LOC_Os07g14850 |
| Insertion | Chr8 | 27430859 | 27430860 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 3523491 | 3554083 | LOC_Os03g06960 |
| Inversion | Chr3 | 3523577 | 3561138 | |
| Inversion | Chr3 | 15783594 | 15795900 | 2 |
| Inversion | Chr3 | 15783606 | 15795900 | 2 |
| Inversion | Chr1 | 22956132 | 23224255 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 22701512 | Chr4 | 28778256 | LOC_Os06g38320 |
