Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN951-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN951-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 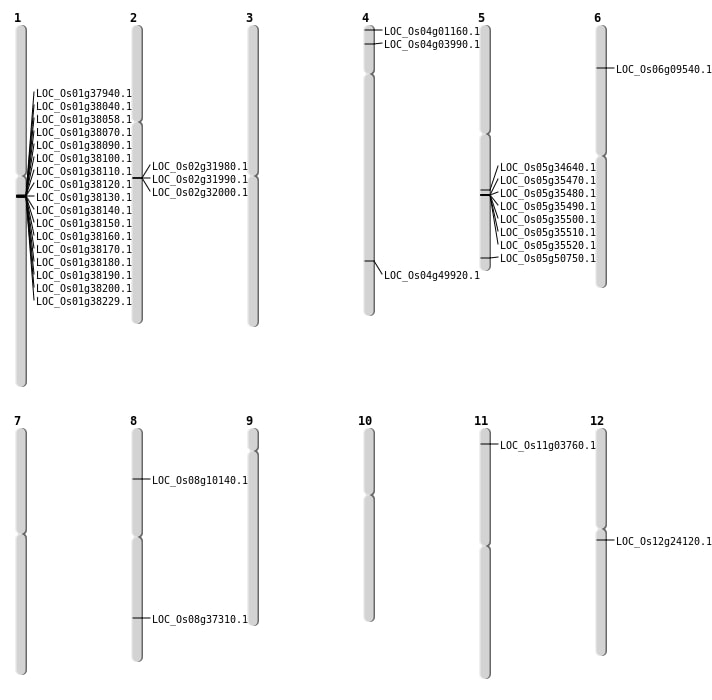
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12659028 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 16375189 | A-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 18843691 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 19569117 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 21254776 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g37940 |
| SBS | Chr1 | 24798578 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 25018099 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 25018100 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 36510792 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 5254442 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 19245918 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 2835352 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 19642123 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 25739626 | C-T | HET | INTRON | |
| SBS | Chr2 | 12149827 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 20065538 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 15945220 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 33672978 | C-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 34087463 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 26470267 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 14029818 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 1907146 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 20543553 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g34640 |
| SBS | Chr5 | 28202319 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr5 | 29097831 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g50750 |
| SBS | Chr5 | 29384407 | A-C | HET | UTR_3_PRIME | |
| SBS | Chr5 | 9053684 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 4681255 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 4681259 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 6284035 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 22676948 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 28600239 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 7975603 | T-C | HET | INTRON | |
| SBS | Chr8 | 13879826 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 5881609 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g10140 |
| SBS | Chr8 | 9463745 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 21977445 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 21977446 | C-T | HET | INTERGENIC |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 123387 | 123389 | 2 | LOC_Os04g01160 |
| Deletion | Chr4 | 1838998 | 1839019 | 21 | LOC_Os04g03990 |
| Deletion | Chr4 | 3412644 | 3412652 | 8 | |
| Deletion | Chr7 | 8209940 | 8209968 | 28 | |
| Deletion | Chr1 | 10976656 | 10976659 | 3 | |
| Deletion | Chr3 | 11816052 | 11816054 | 2 | |
| Deletion | Chr2 | 11987552 | 11987563 | 11 | |
| Deletion | Chr12 | 13724960 | 13724961 | 1 | LOC_Os12g24120 |
| Deletion | Chr5 | 13863292 | 13863295 | 3 | |
| Deletion | Chr6 | 15555237 | 15555241 | 4 | |
| Deletion | Chr6 | 15720339 | 15720342 | 3 | |
| Deletion | Chr3 | 17835643 | 17835652 | 9 | |
| Deletion | Chr2 | 18902333 | 18906015 | 3682 | 3 |
| Deletion | Chr8 | 19461253 | 19461280 | 27 | |
| Deletion | Chr5 | 21079001 | 21120000 | 41000 | 6 |
| Deletion | Chr1 | 21311001 | 21428000 | 117000 | 16 |
| Deletion | Chr8 | 24994811 | 24994812 | 1 | |
| Deletion | Chr3 | 25079997 | 25079998 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr7 | 12555072 | 12555073 | 2 | |
| Insertion | Chr8 | 25824153 | 25824154 | 2 | |
| Insertion | Chr8 | 8657538 | 8657538 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 969301 | 969439 | |
| Inversion | Chr11 | 9134393 | 27265439 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1474536 | Chr10 | 19565345 | LOC_Os11g03760 |
| Translocation | Chr11 | 1475413 | Chr10 | 19565327 | LOC_Os11g03760 |
| Translocation | Chr6 | 4846178 | Chr5 | 21119986 | LOC_Os06g09540 |
| Translocation | Chr8 | 21008272 | Chr4 | 29771154 | LOC_Os04g49920 |
| Translocation | Chr8 | 23577081 | Chr6 | 4841834 | LOC_Os08g37310 |
| Translocation | Chr8 | 23577093 | Chr5 | 21120312 | 2 |
