Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN955-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN955-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 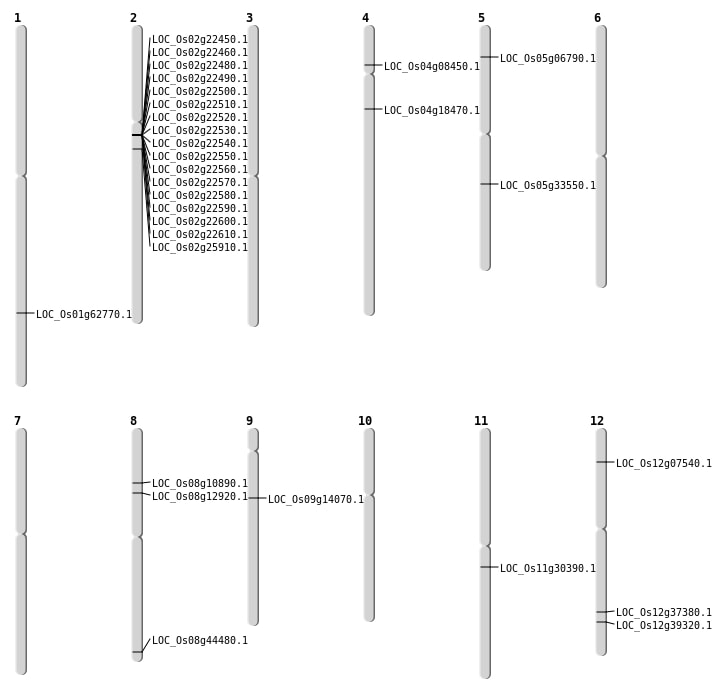
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17721288 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 27316439 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 3947511 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 40789897 | T-A | HET | INTRON | |
| SBS | Chr1 | 41027212 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 10006203 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 26623013 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 17452361 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 3756811 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g07540 |
| SBS | Chr2 | 14750482 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 14750483 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 19400561 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 3983349 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 11145479 | T-A | HET | INTRON | |
| SBS | Chr5 | 18108885 | A-G | HOMO | INTRON | |
| SBS | Chr5 | 18125496 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 9684657 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 16963928 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 28706966 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 27973245 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g44480 |
| SBS | Chr8 | 28427156 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 3716198 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 15989219 | G-T | HOMO | INTRON | |
| SBS | Chr9 | 3969132 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 8330241 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g14070 |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2851931 | 2851932 | 1 | |
| Deletion | Chr6 | 2853138 | 2853141 | 3 | |
| Deletion | Chr10 | 3544149 | 3544158 | 9 | |
| Deletion | Chr3 | 3930185 | 3930186 | 1 | |
| Deletion | Chr8 | 6420119 | 6420120 | 1 | LOC_Os08g10890 |
| Deletion | Chr8 | 8409544 | 8409545 | 1 | |
| Deletion | Chr1 | 10098199 | 10098202 | 3 | |
| Deletion | Chr2 | 12854366 | 12855402 | 1036 | |
| Deletion | Chr2 | 13408728 | 13484721 | 75993 | 16 |
| Deletion | Chr1 | 14567120 | 14567126 | 6 | |
| Deletion | Chr11 | 17643001 | 17662000 | 19000 | LOC_Os11g30390 |
| Deletion | Chr9 | 17819352 | 17819357 | 5 | |
| Deletion | Chr7 | 17880641 | 17880659 | 18 | |
| Deletion | Chr2 | 18063988 | 18063995 | 7 | |
| Deletion | Chr10 | 22367937 | 22367945 | 8 | |
| Deletion | Chr1 | 24898726 | 24898731 | 5 | |
| Deletion | Chr6 | 27198821 | 27198827 | 6 | |
| Deletion | Chr7 | 28222400 | 28222407 | 7 | |
| Deletion | Chr5 | 29058489 | 29058541 | 52 | |
| Deletion | Chr3 | 33859075 | 33859076 | 1 | |
| Deletion | Chr1 | 36347592 | 36347593 | 1 | LOC_Os01g62770 |
Insertions: 6
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 5140634 | 19704713 | LOC_Os05g33550 |
| Inversion | Chr8 | 7681507 | 8484377 | LOC_Os08g12920 |
| Inversion | Chr8 | 12951566 | 12984920 | |
| Inversion | Chr2 | 13408740 | 14727717 | |
| Inversion | Chr2 | 14727714 | 15206418 | LOC_Os02g25910 |
| Inversion | Chr11 | 17643535 | 18979933 | |
| Inversion | Chr12 | 22949880 | 24195206 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3129575 | Chr6 | 18189892 | |
| Translocation | Chr5 | 3546434 | Chr3 | 22228349 | LOC_Os05g06790 |
| Translocation | Chr4 | 4546449 | Chr2 | 12854381 | LOC_Os04g08450 |
| Translocation | Chr11 | 13259842 | Chr4 | 10208284 | LOC_Os04g18470 |
| Translocation | Chr5 | 19704713 | Chr3 | 30857701 | LOC_Os05g33550 |
| Translocation | Chr5 | 19704895 | Chr3 | 30859398 | LOC_Os05g33550 |
| Translocation | Chr8 | 27508617 | Chr2 | 12855389 | |
| Translocation | Chr8 | 27508622 | Chr2 | 12854731 |
