Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN957-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN957-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 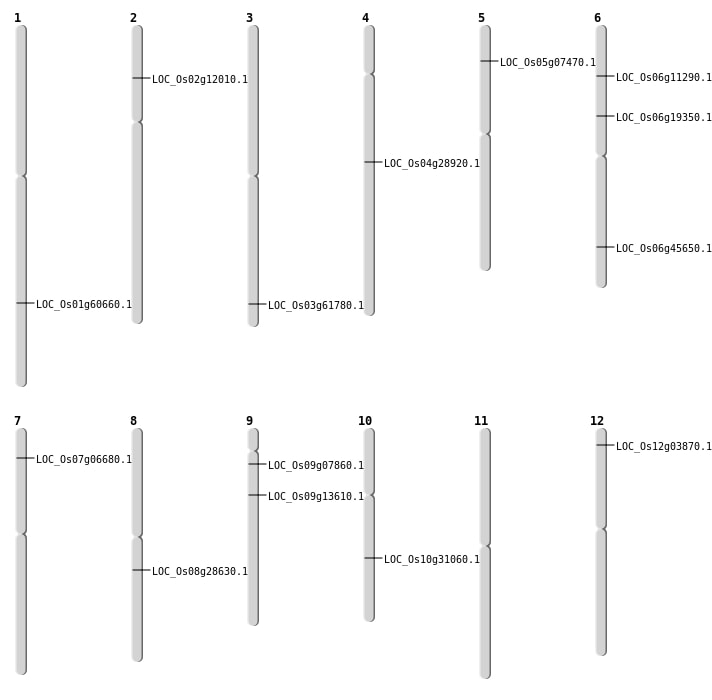
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20696672 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 24292437 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 35072332 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g60660 |
| SBS | Chr1 | 37437871 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 12654074 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 13909150 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 6884947 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 6884948 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 6884950 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11454483 | T-C | HET | INTRON | |
| SBS | Chr11 | 13354582 | C-A | HOMO | INTRON | |
| SBS | Chr12 | 1595746 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g03870 |
| SBS | Chr12 | 1595747 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g03870 |
| SBS | Chr12 | 21541395 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 10345265 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 20682265 | A-C | HET | UTR_3_PRIME | |
| SBS | Chr2 | 27609799 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 24786651 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 30157618 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 7129959 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 9094057 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 17144231 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g28920 |
| SBS | Chr4 | 33349278 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 12804950 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 1965199 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 20613439 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 3982434 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g07470 |
| SBS | Chr5 | 4815373 | G-A | HOMO | UTR_5_PRIME | |
| SBS | Chr6 | 20243509 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 30158126 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 18165406 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 26844200 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 26963023 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 3442882 | G-A | HET | INTRON | |
| SBS | Chr7 | 8563563 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 18836910 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 27018798 | C-T | HET | INTRON | |
| SBS | Chr9 | 10518865 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 18689160 | T-A | HET | INTRON | |
| SBS | Chr9 | 3997712 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g07860 |
| SBS | Chr9 | 7920849 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g13610 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 562001 | 562010 | 9 | |
| Deletion | Chr7 | 3259680 | 3259695 | 15 | LOC_Os07g06680 |
| Deletion | Chr5 | 3441786 | 3441797 | 11 | |
| Deletion | Chr8 | 5587919 | 5587920 | 1 | |
| Deletion | Chr6 | 5918705 | 5918711 | 6 | LOC_Os06g11290 |
| Deletion | Chr2 | 6229592 | 6229605 | 13 | LOC_Os02g12010 |
| Deletion | Chr8 | 8427612 | 8427615 | 3 | |
| Deletion | Chr4 | 9402670 | 9402673 | 3 | |
| Deletion | Chr8 | 10033778 | 10033781 | 3 | |
| Deletion | Chr12 | 10311384 | 10311397 | 13 | |
| Deletion | Chr11 | 14067621 | 14067631 | 10 | |
| Deletion | Chr10 | 16247064 | 16247065 | 1 | LOC_Os10g31060 |
| Deletion | Chr5 | 17885343 | 17885344 | 1 | |
| Deletion | Chr3 | 21595408 | 21595415 | 7 | |
| Deletion | Chr4 | 22897318 | 22897331 | 13 | |
| Deletion | Chr7 | 25263695 | 25263699 | 4 | |
| Deletion | Chr2 | 27145056 | 27145060 | 4 | |
| Deletion | Chr6 | 27639070 | 27639071 | 1 | LOC_Os06g45650 |
| Deletion | Chr2 | 32333331 | 32333332 | 1 | |
| Deletion | Chr3 | 35028498 | 35028601 | 103 | LOC_Os03g61780 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 18552323 | 18552324 | 2 | |
| Insertion | Chr4 | 7471318 | 7471318 | 1 | |
| Insertion | Chr5 | 29627689 | 29627692 | 4 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 24346819 | 41519862 | |
| Inversion | Chr3 | 24396561 | 24958769 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 11010401 | Chr4 | 11634136 | LOC_Os06g19350 |
| Translocation | Chr4 | 15896407 | Chr1 | 9105546 | |
| Translocation | Chr4 | 19818386 | Chr2 | 28876935 | |
| Translocation | Chr11 | 21092273 | Chr8 | 17496660 | LOC_Os08g28630 |
| Translocation | Chr11 | 27827916 | Chr7 | 17812786 |
