Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN959-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN959-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 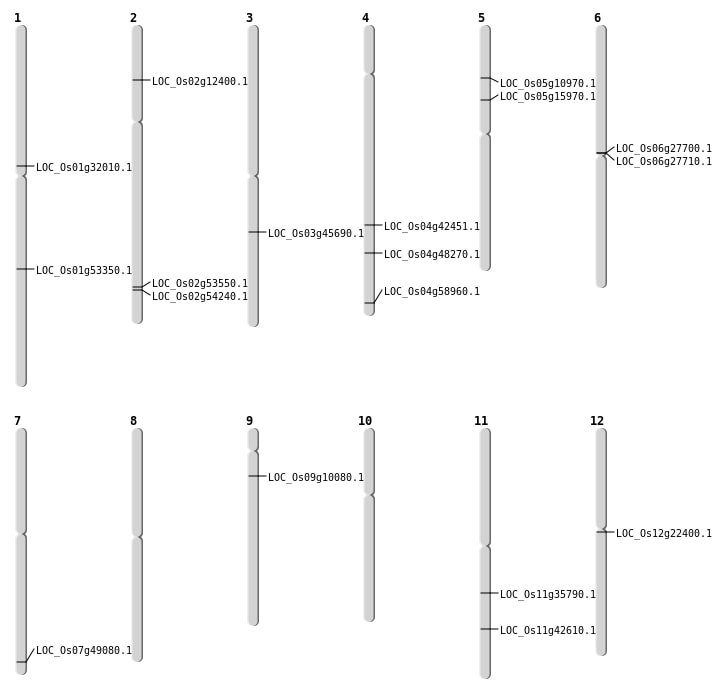
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10609653 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 13334067 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 17529841 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g32010 |
| SBS | Chr1 | 30656297 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g53350 |
| SBS | Chr1 | 30656298 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 40766934 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr1 | 41526720 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 5486531 | T-C | HOMO | INTRON | |
| SBS | Chr1 | 5486599 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 11563215 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 12561329 | A-T | HET | INTRON | |
| SBS | Chr10 | 12561330 | C-T | HET | INTRON | |
| SBS | Chr10 | 21360027 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr11 | 11477660 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 21010216 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g35790 |
| SBS | Chr12 | 10271759 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 12651513 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g22400 |
| SBS | Chr12 | 5296102 | C-A | HOMO | INTRON | |
| SBS | Chr2 | 32764606 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g53550 |
| SBS | Chr2 | 6469408 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g12400 |
| SBS | Chr2 | 6469409 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g12400 |
| SBS | Chr3 | 17296025 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 22937377 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 10957882 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 12702137 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 21809983 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 21809984 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 33076747 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 20509306 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 28489190 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 9017603 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15970 |
| SBS | Chr6 | 24183411 | T-A | HOMO | INTRON | |
| SBS | Chr6 | 26041573 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 12544540 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 25797079 | C-G | HOMO | UTR_3_PRIME | |
| SBS | Chr7 | 28856528 | A-T | HET | INTRON | |
| SBS | Chr7 | 3353902 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 7898453 | A-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 14756658 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 18945219 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 5495450 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2737703 | 2737716 | 13 | |
| Deletion | Chr7 | 5592044 | 5592056 | 12 | |
| Deletion | Chr10 | 5740834 | 5740841 | 7 | |
| Deletion | Chr4 | 8060808 | 8060815 | 7 | |
| Deletion | Chr7 | 10467936 | 10467937 | 1 | |
| Deletion | Chr3 | 11311144 | 11311153 | 9 | |
| Deletion | Chr6 | 12843378 | 12843385 | 7 | |
| Deletion | Chr6 | 15683188 | 15688412 | 5224 | 2 |
| Deletion | Chr3 | 17198748 | 17198751 | 3 | |
| Deletion | Chr12 | 23059125 | 23059134 | 9 | |
| Deletion | Chr3 | 24310897 | 24310905 | 8 | |
| Deletion | Chr4 | 25122546 | 25122549 | 3 | LOC_Os04g42451 |
| Deletion | Chr5 | 25273810 | 25273816 | 6 | |
| Deletion | Chr1 | 26927426 | 26927431 | 5 | |
| Deletion | Chr4 | 28754165 | 28754170 | 5 | LOC_Os04g48270 |
| Deletion | Chr7 | 29382934 | 29383160 | 226 | LOC_Os07g49080 |
| Deletion | Chr2 | 33250184 | 33250201 | 17 | LOC_Os02g54240 |
| Deletion | Chr1 | 34286304 | 34286346 | 42 | |
| Deletion | Chr4 | 35074674 | 35074677 | 3 | LOC_Os04g58960 |
| Deletion | Chr1 | 40770942 | 40770943 | 1 |
Insertions: 7
Inversions: 5
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 6127528 | Chr3 | 25791631 | 2 |
| Translocation | Chr12 | 8904621 | Chr10 | 18963096 | |
| Translocation | Chr8 | 15269709 | Chr2 | 12182624 | |
| Translocation | Chr3 | 25354096 | Chr1 | 27882664 | |
| Translocation | Chr11 | 25650484 | Chr7 | 9569163 | LOC_Os11g42610 |
