Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN961-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN961-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 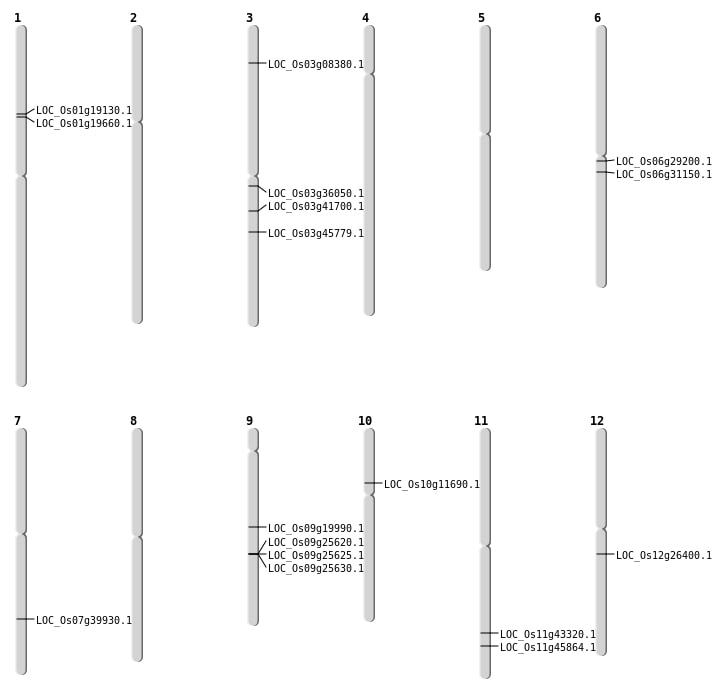
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10799846 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 10805795 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g19130 |
| SBS | Chr1 | 13655503 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 275870 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 29114553 | C-T | HET | INTRON | |
| SBS | Chr1 | 30237907 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 30237909 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 6282139 | G-C | HET | INTRON | |
| SBS | Chr10 | 15468601 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 6460145 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g11690 |
| SBS | Chr11 | 11314293 | A-T | HOMO | INTRON | |
| SBS | Chr12 | 11667645 | A-G | HOMO | INTRON | |
| SBS | Chr12 | 15427805 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g26400 |
| SBS | Chr2 | 26304188 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 19996267 | C-T | HET | STOP_GAINED | LOC_Os03g36050 |
| SBS | Chr3 | 25860663 | G-A | HET | START_GAINED | LOC_Os03g45779 |
| SBS | Chr3 | 31973448 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 8880081 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 10066927 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 10300837 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 16931935 | T-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr4 | 23624265 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 28706548 | A-G | HOMO | INTRON | |
| SBS | Chr4 | 5622410 | C-T | HET | INTRON | |
| SBS | Chr4 | 5979877 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 15929375 | G-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 26112024 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 5104850 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 14033230 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 18096316 | C-T | HET | SPLICE_SITE_DONOR | LOC_Os06g31150 |
| SBS | Chr6 | 24407498 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 10579040 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19044128 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 19156138 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 23954697 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g39930 |
| SBS | Chr7 | 23995263 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 25584515 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 25584516 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 513427 | G-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 8807498 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 9188721 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 11786831 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 12523399 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 7048031 | G-A | HOMO | INTRON |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 935210 | 935212 | 2 | |
| Deletion | Chr3 | 4280653 | 4280663 | 10 | LOC_Os03g08380 |
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr4 | 5032756 | 5032760 | 4 | |
| Deletion | Chr2 | 5841761 | 5841780 | 19 | |
| Deletion | Chr8 | 6607748 | 6607749 | 1 | |
| Deletion | Chr4 | 6736859 | 6736876 | 17 | |
| Deletion | Chr1 | 8699044 | 8699045 | 1 | |
| Deletion | Chr6 | 10486890 | 10486891 | 1 | |
| Deletion | Chr11 | 11091317 | 11091320 | 3 | |
| Deletion | Chr1 | 11148749 | 11148755 | 6 | LOC_Os01g19660 |
| Deletion | Chr5 | 14003433 | 14003443 | 10 | |
| Deletion | Chr9 | 15386001 | 15397000 | 11000 | 3 |
| Deletion | Chr9 | 15481301 | 15481302 | 1 | |
| Deletion | Chr1 | 17859116 | 17859117 | 1 | |
| Deletion | Chr5 | 18706226 | 18706251 | 25 | |
| Deletion | Chr6 | 21097154 | 21097171 | 17 | |
| Deletion | Chr3 | 23168643 | 23168655 | 12 | LOC_Os03g41700 |
| Deletion | Chr3 | 26175512 | 26175534 | 22 | |
| Deletion | Chr1 | 26447338 | 26447339 | 1 | |
| Deletion | Chr11 | 26618591 | 26618592 | 1 | |
| Deletion | Chr7 | 27652662 | 27652663 | 1 | |
| Deletion | Chr1 | 32133882 | 32133905 | 23 | |
| Deletion | Chr4 | 34310159 | 34310161 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23445831 | 23445832 | 2 | |
| Insertion | Chr10 | 14914819 | 14914819 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 15385579 | 15400051 | |
| Inversion | Chr9 | 15396845 | 15400079 | |
| Inversion | Chr11 | 26141370 | 26174683 | LOC_Os11g43320 |
| Inversion | Chr11 | 26141394 | 26174690 | LOC_Os11g43320 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 16666430 | Chr1 | 11109755 | LOC_Os06g29200 |
| Translocation | Chr8 | 16788238 | Chr3 | 19478448 | |
| Translocation | Chr11 | 27744306 | Chr9 | 11970466 | 2 |
