Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN978-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN978-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 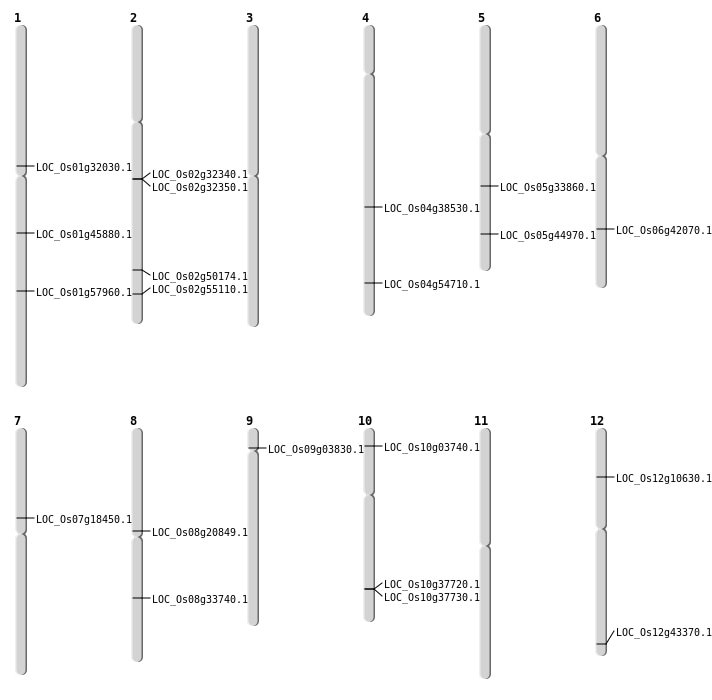
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10567248 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 22256596 | T-A | HET | INTRON | |
| SBS | Chr1 | 7191280 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 1691227 | G-A | HET | START_GAINED | LOC_Os10g03740 |
| SBS | Chr10 | 18941317 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr11 | 17689817 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 24262431 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 18211695 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 1898014 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 277670 | G-T | HET | INTRON | |
| SBS | Chr2 | 19101526 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g32350 |
| SBS | Chr2 | 30652947 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g50174 |
| SBS | Chr2 | 30654846 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 31696381 | C-A | HET | INTRON | |
| SBS | Chr2 | 32227242 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 34547853 | T-C | HET | INTRON | |
| SBS | Chr2 | 4108019 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 7817220 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 16340082 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 17233482 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 19287231 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 32364484 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 33349580 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 33349581 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 32540853 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g54710 |
| SBS | Chr4 | 8979393 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 2022903 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 11646889 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 16172062 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 16172063 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 20411993 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 24615311 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 27955752 | C-T | HET | INTRON | |
| SBS | Chr6 | 4535826 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 22588144 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 25751570 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 3740678 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 5951715 | C-T | HET | INTRON | |
| SBS | Chr7 | 7172121 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 12508393 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g20849 |
| SBS | Chr9 | 15510050 | A-G | HOMO | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1750369 | 1750377 | 8 | |
| Deletion | Chr4 | 2390839 | 2390846 | 7 | |
| Deletion | Chr6 | 4564810 | 4564815 | 5 | |
| Deletion | Chr1 | 5007522 | 5007533 | 11 | |
| Deletion | Chr1 | 5104394 | 5104401 | 7 | |
| Deletion | Chr12 | 5672519 | 5674643 | 2124 | LOC_Os12g10630 |
| Deletion | Chr4 | 10220906 | 10220917 | 11 | |
| Deletion | Chr7 | 10912926 | 10912930 | 4 | LOC_Os07g18450 |
| Deletion | Chr2 | 12298549 | 12298575 | 26 | |
| Deletion | Chr8 | 14907148 | 14907156 | 8 | |
| Deletion | Chr9 | 15585045 | 15585046 | 1 | |
| Deletion | Chr3 | 16207521 | 16207538 | 17 | |
| Deletion | Chr3 | 16411894 | 16411904 | 10 | |
| Deletion | Chr2 | 19091412 | 19091434 | 22 | LOC_Os02g32340 |
| Deletion | Chr11 | 19191736 | 19191739 | 3 | |
| Deletion | Chr9 | 19593456 | 19593457 | 1 | |
| Deletion | Chr5 | 19978072 | 19978083 | 11 | LOC_Os05g33860 |
| Deletion | Chr10 | 20185329 | 20185349 | 20 | 2 |
| Deletion | Chr1 | 20907223 | 20907233 | 10 | |
| Deletion | Chr5 | 23498148 | 23498153 | 5 | |
| Deletion | Chr3 | 25195037 | 25195038 | 1 | |
| Deletion | Chr5 | 25511379 | 25511441 | 62 | |
| Deletion | Chr5 | 26146170 | 26146173 | 3 | LOC_Os05g44970 |
| Deletion | Chr12 | 26266653 | 26266660 | 7 | |
| Deletion | Chr2 | 32513468 | 32513487 | 19 | |
| Deletion | Chr2 | 33758045 | 33758048 | 3 | LOC_Os02g55110 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2329880 | 2329881 | 2 | |
| Insertion | Chr12 | 26909980 | 26909982 | 3 | LOC_Os12g43370 |
| Insertion | Chr2 | 28012238 | 28012239 | 2 | |
| Insertion | Chr4 | 312993 | 312994 | 2 | |
| Insertion | Chr6 | 14639836 | 14639836 | 1 | |
| Insertion | Chr6 | 7477529 | 7477529 | 1 |
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 7229234 | 33506545 | LOC_Os01g57960 |
| Inversion | Chr1 | 7375554 | 7804133 | |
| Inversion | Chr1 | 7375567 | 7804156 | |
| Inversion | Chr1 | 10567471 | 11325213 | |
| Inversion | Chr1 | 17167774 | 17535659 | LOC_Os01g32030 |
| Inversion | Chr8 | 20785819 | 21078843 | LOC_Os08g33740 |
| Inversion | Chr8 | 20785830 | 21078856 | LOC_Os08g33740 |
| Inversion | Chr3 | 28924665 | 29073843 | |
| Inversion | Chr3 | 28946192 | 29073843 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1943435 | Chr6 | 25273419 | 2 |
| Translocation | Chr5 | 3543877 | Chr3 | 13563773 | |
| Translocation | Chr2 | 13467141 | Chr1 | 26060845 | LOC_Os01g45880 |
| Translocation | Chr10 | 18628322 | Chr5 | 9638214 | |
| Translocation | Chr12 | 20909879 | Chr6 | 8520812 | |
| Translocation | Chr4 | 22896294 | Chr2 | 13396941 | LOC_Os04g38530 |
| Translocation | Chr3 | 28946283 | Chr2 | 21947216 |
