Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | W138-2-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download W138-2-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 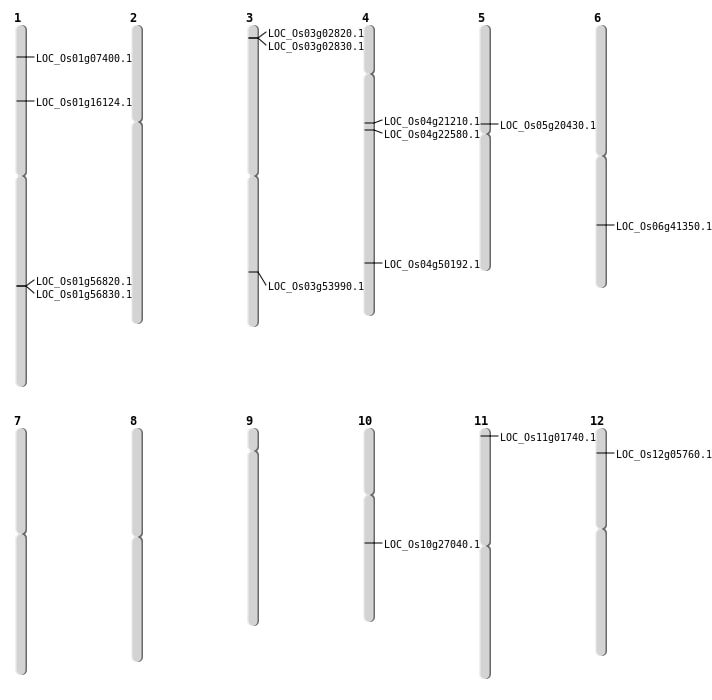
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13433954 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 14258265 | T-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 22019656 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 32283556 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 42722116 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 7554676 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 14251949 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g27040 |
| SBS | Chr10 | 16077566 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 16221895 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 18096746 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 21611568 | A-G | HET | INTRON | |
| SBS | Chr11 | 12973316 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 14335540 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr11 | 17260325 | C-G | HET | INTRON | |
| SBS | Chr11 | 17944292 | G-T | HOMO | INTRON | |
| SBS | Chr11 | 8047233 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 11865316 | T-C | HET | INTRON | |
| SBS | Chr12 | 26828711 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 5181001 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 7498583 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 13290912 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 13987366 | A-G | HOMO | UTR_5_PRIME | |
| SBS | Chr3 | 12830027 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 24567364 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 27964385 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 7338112 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 11959781 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g21210 |
| SBS | Chr4 | 12783441 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g22580 |
| SBS | Chr4 | 29949417 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g50192 |
| SBS | Chr4 | 33457388 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 4935401 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 11988951 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g20430 |
| SBS | Chr5 | 28700202 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 30871799 | C-T | HET | INTRON | |
| SBS | Chr7 | 14308740 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19106180 | G-A | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1102837 | 1102845 | 8 | 2 |
| Deletion | Chr7 | 1220218 | 1220219 | 1 | |
| Deletion | Chr12 | 2653530 | 2653531 | 1 | LOC_Os12g05760 |
| Deletion | Chr8 | 2795408 | 2795416 | 8 | |
| Deletion | Chr3 | 4583082 | 4583083 | 1 | |
| Deletion | Chr1 | 9099151 | 9099156 | 5 | LOC_Os01g16124 |
| Deletion | Chr10 | 11012459 | 11012469 | 10 | |
| Deletion | Chr5 | 11251687 | 11251689 | 2 | |
| Deletion | Chr9 | 12814382 | 12814384 | 2 | |
| Deletion | Chr7 | 13170460 | 13170569 | 109 | |
| Deletion | Chr6 | 14761307 | 14761308 | 1 | |
| Deletion | Chr4 | 15085746 | 15085762 | 16 | |
| Deletion | Chr2 | 15966632 | 15966636 | 4 | |
| Deletion | Chr6 | 17499979 | 17499982 | 3 | |
| Deletion | Chr3 | 18281808 | 18281820 | 12 | |
| Deletion | Chr2 | 18401974 | 18401986 | 12 | |
| Deletion | Chr12 | 20050528 | 20050529 | 1 | |
| Deletion | Chr1 | 21521012 | 21521013 | 1 | |
| Deletion | Chr1 | 21734829 | 21734847 | 18 | |
| Deletion | Chr6 | 24769168 | 24769169 | 1 | LOC_Os06g41350 |
| Deletion | Chr5 | 25710988 | 25712503 | 1515 | |
| Deletion | Chr2 | 30443419 | 30443420 | 1 | |
| Deletion | Chr1 | 32812551 | 32825919 | 13368 | 2 |
| Deletion | Chr4 | 33455020 | 33455032 | 12 | |
| Deletion | Chr1 | 34217154 | 34217155 | 1 |
Insertions: 5
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 28794914 | 30957515 | LOC_Os03g53990 |
| Inversion | Chr3 | 28794927 | 30957524 | LOC_Os03g53990 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 399375 | Chr1 | 32812553 | LOC_Os11g01740 |
| Translocation | Chr11 | 399380 | Chr1 | 32825925 | LOC_Os11g01740 |
| Translocation | Chr12 | 9761058 | Chr1 | 3507037 | LOC_Os01g07400 |
| Translocation | Chr12 | 9800967 | Chr1 | 3507049 | LOC_Os01g07400 |
| Translocation | Chr11 | 10905164 | Chr5 | 25710995 |
