Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | W247-2-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download W247-2-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 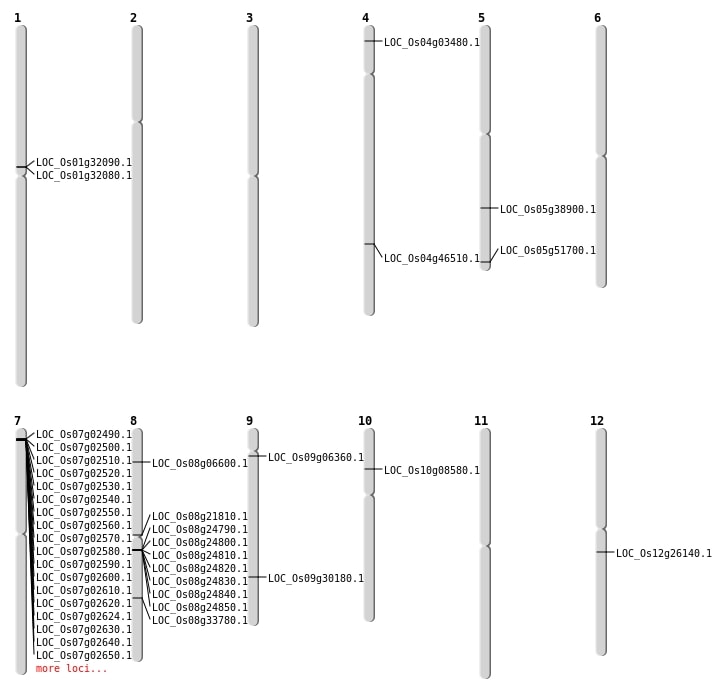
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 187430 | A-T | HET | INTRON | |
| SBS | Chr1 | 21442377 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 34530187 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 12526039 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 13917058 | A-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 421475 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 15225739 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g26140 |
| SBS | Chr12 | 26562552 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 11644067 | G-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 15474068 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 19471436 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 26549320 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 31451198 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 31550992 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 3506851 | G-A | HET | INTRON | |
| SBS | Chr4 | 18189650 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 22208180 | T-G | HET | INTRON | |
| SBS | Chr4 | 27587980 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g46510 |
| SBS | Chr4 | 5980377 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 7222760 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 15705919 | G-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 21239966 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 2368620 | G-A | HET | INTRON | |
| SBS | Chr5 | 29658542 | G-T | HET | INTRON | |
| SBS | Chr6 | 9314987 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 10646502 | G-A | HET | INTRON | |
| SBS | Chr7 | 19798629 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g33150 |
| SBS | Chr7 | 25052934 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g41830 |
| SBS | Chr7 | 67670 | G-A | HOMO | UTR_5_PRIME | |
| SBS | Chr8 | 20132041 | C-G | HOMO | INTRON | |
| SBS | Chr8 | 21127616 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g33780 |
| SBS | Chr8 | 21331647 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 3725989 | G-A | HET | INTRON | |
| SBS | Chr9 | 2992485 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g06360 |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 163566 | 163569 | 3 | |
| Deletion | Chr7 | 864001 | 1023000 | 159000 | 30 |
| Deletion | Chr4 | 1476313 | 1476314 | 1 | |
| Deletion | Chr4 | 1504916 | 1506057 | 1141 | LOC_Os04g03480 |
| Deletion | Chr8 | 3726085 | 3726088 | 3 | LOC_Os08g06600 |
| Deletion | Chr11 | 4687166 | 4687189 | 23 | |
| Deletion | Chr6 | 5563444 | 5563446 | 2 | |
| Deletion | Chr7 | 7767148 | 7767156 | 8 | |
| Deletion | Chr10 | 9066263 | 9066264 | 1 | |
| Deletion | Chr12 | 11126894 | 11126899 | 5 | |
| Deletion | Chr10 | 12191565 | 12191583 | 18 | |
| Deletion | Chr9 | 12434585 | 12434586 | 1 | |
| Deletion | Chr8 | 13057462 | 13057463 | 1 | LOC_Os08g21810 |
| Deletion | Chr1 | 13643952 | 13643957 | 5 | |
| Deletion | Chr5 | 13959517 | 13959521 | 4 | |
| Deletion | Chr8 | 15008001 | 15040000 | 32000 | 7 |
| Deletion | Chr7 | 17489324 | 17489327 | 3 | |
| Deletion | Chr1 | 17564474 | 17564477 | 3 | 2 |
| Deletion | Chr3 | 23690063 | 23690064 | 1 | |
| Deletion | Chr2 | 25900068 | 25900071 | 3 | |
| Deletion | Chr8 | 26491772 | 26492271 | 499 | |
| Deletion | Chr5 | 29661281 | 29661325 | 44 | LOC_Os05g51700 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 3254413 | 3254414 | 2 | |
| Insertion | Chr6 | 22140214 | 22140215 | 2 | |
| Insertion | Chr7 | 28510090 | 28510090 | 1 | |
| Insertion | Chr9 | 18368026 | 18368026 | 1 | LOC_Os09g30180 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 21254552 | 21728807 | |
| Inversion | Chr2 | 22892196 | 23473294 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 1551044 | Chr5 | 22817095 | LOC_Os05g38900 |
| Translocation | Chr10 | 4663965 | Chr4 | 1504927 | LOC_Os10g08580 |
| Translocation | Chr12 | 14725326 | Chr4 | 1506053 | LOC_Os04g03480 |
| Translocation | Chr12 | 14728112 | Chr10 | 4663971 | LOC_Os10g08580 |
| Translocation | Chr8 | 26491767 | Chr5 | 22817054 | LOC_Os05g38900 |
| Translocation | Chr8 | 26492263 | Chr7 | 1022873 |
