Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | W423-5-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download W423-5-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 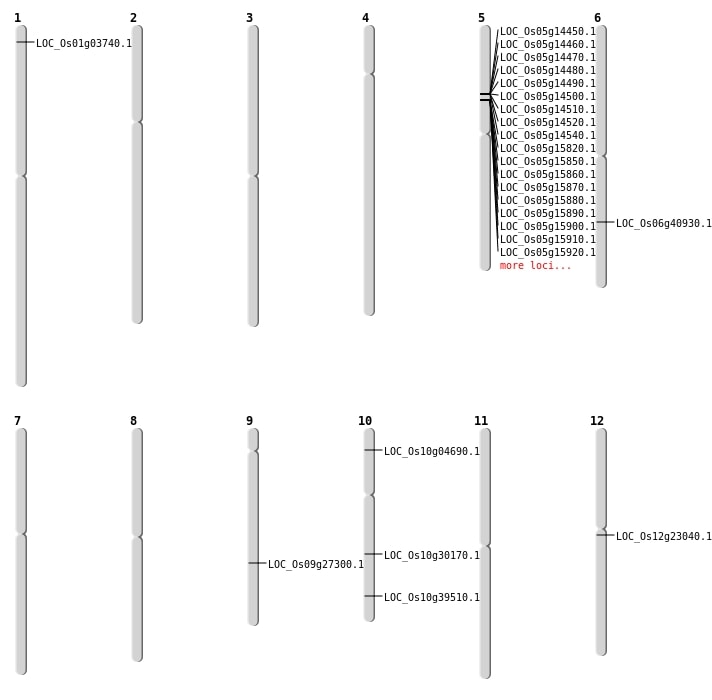
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15821009 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 28239815 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 3191504 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 3191505 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 21601025 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 3603423 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 770230 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 24338477 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 13026237 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g23040 |
| SBS | Chr12 | 18439224 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 20279227 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 1962463 | A-G | HET | INTRON | |
| SBS | Chr2 | 8692416 | A-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 11450939 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 18013777 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 23006070 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 3891839 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 7973906 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 17054331 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 17676636 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 11018639 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 13872351 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 26962197 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 4627738 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 4847597 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 8247051 | T-A | HET | INTRON | |
| SBS | Chr5 | 8354855 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 10443736 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 24396629 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g40930 |
| SBS | Chr7 | 29222337 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 2278886 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 16600390 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g27300 |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1564742 | 1564744 | 2 | LOC_Os01g03740 |
| Deletion | Chr3 | 5183272 | 5183346 | 74 | |
| Deletion | Chr3 | 5472881 | 5472886 | 5 | |
| Deletion | Chr3 | 7951797 | 7951801 | 4 | |
| Deletion | Chr5 | 8155001 | 8247000 | 92000 | 9 |
| Deletion | Chr5 | 8370907 | 8370912 | 5 | |
| Deletion | Chr9 | 8420136 | 8420137 | 1 | |
| Deletion | Chr12 | 8739246 | 8739251 | 5 | |
| Deletion | Chr8 | 8920338 | 8920354 | 16 | |
| Deletion | Chr5 | 8934001 | 9112000 | 178000 | 28 |
| Deletion | Chr3 | 9698146 | 9698148 | 2 | |
| Deletion | Chr5 | 10310001 | 10352000 | 42000 | 8 |
| Deletion | Chr10 | 11081467 | 11081472 | 5 | |
| Deletion | Chr6 | 12328336 | 12328338 | 2 | |
| Deletion | Chr12 | 13184718 | 13184725 | 7 | |
| Deletion | Chr4 | 14229807 | 14229809 | 2 | |
| Deletion | Chr10 | 15684246 | 15684250 | 4 | LOC_Os10g30170 |
| Deletion | Chr10 | 16899083 | 16899085 | 2 | |
| Deletion | Chr10 | 17673113 | 17673115 | 2 | |
| Deletion | Chr2 | 17755097 | 17755098 | 1 | |
| Deletion | Chr7 | 24701433 | 24701434 | 1 | |
| Deletion | Chr6 | 26781856 | 26781858 | 2 | |
| Deletion | Chr3 | 27033715 | 27041247 | 7532 | |
| Deletion | Chr6 | 27085904 | 27085905 | 1 | |
| Deletion | Chr1 | 29838836 | 29838840 | 4 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2743564 | 2743566 | 3 | |
| Insertion | Chr5 | 4809099 | 4809101 | 3 | |
| Insertion | Chr6 | 19413111 | 19413112 | 2 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 1458192 | 2237759 | |
| Inversion | Chr10 | 1458211 | 2240769 | LOC_Os10g04690 |
| Inversion | Chr5 | 8155064 | 13511514 | |
| Inversion | Chr5 | 8246819 | 13511517 | LOC_Os05g14540 |
| Inversion | Chr12 | 18081479 | 18083626 | |
| Inversion | Chr12 | 18081492 | 18083631 | |
| Inversion | Chr2 | 27440261 | 27748536 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2237774 | Chr5 | 10309838 | LOC_Os05g17930 |
| Translocation | Chr10 | 2240749 | Chr4 | 2491099 | LOC_Os10g04690 |
| Translocation | Chr12 | 3653300 | Chr9 | 6641917 | |
| Translocation | Chr11 | 15322806 | Chr2 | 27748529 | |
| Translocation | Chr11 | 15353170 | Chr3 | 22530088 | |
| Translocation | Chr10 | 21106398 | Chr4 | 14889178 | LOC_Os10g39510 |
| Translocation | Chr3 | 22531418 | Chr2 | 27440253 |
