Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | W470-2-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download W470-2-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 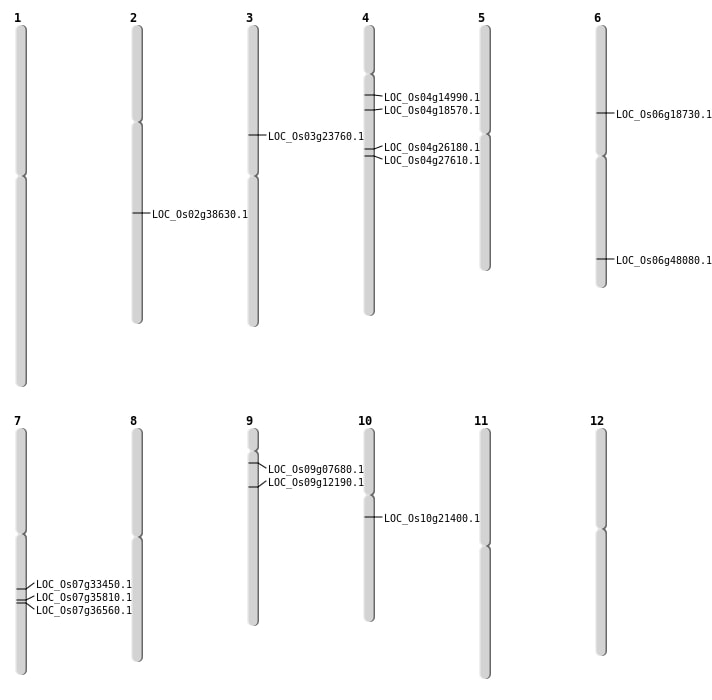
|
| Variant Information |
|---|
Single base substitutions: 63
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13763426 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 20139984 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 29344350 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 41534780 | G-A | HOMO | INTRON | |
| SBS | Chr1 | 42671776 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 7835814 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 18164434 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 8146251 | C-T | HET | INTRON | |
| SBS | Chr12 | 11983875 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 21774717 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 23406393 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 13704278 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 23348223 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g38630 |
| SBS | Chr2 | 31547149 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 31547225 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 4847279 | T-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 5394698 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 7794668 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 7794717 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 13468724 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g23760 |
| SBS | Chr3 | 19567133 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 32195115 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 33119600 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 4034753 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 487107 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 15265140 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g26180 |
| SBS | Chr4 | 15268784 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 16395613 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 24999518 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 2950394 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 35370524 | T-C | HET | INTRON | |
| SBS | Chr4 | 4527400 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 8422541 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g14990 |
| SBS | Chr5 | 11519554 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 1165105 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 1301611 | C-A | HET | INTRON | |
| SBS | Chr5 | 2601687 | A-C | HET | INTRON | |
| SBS | Chr5 | 26929223 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 29956876 | T-C | HET | INTRON | |
| SBS | Chr5 | 3387949 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 9847410 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 10620041 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g18730 |
| SBS | Chr6 | 1815869 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 19595597 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 27682383 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 29094575 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g48080 |
| SBS | Chr6 | 30436203 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 7483762 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 12421380 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 20783800 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 21444090 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g35810 |
| SBS | Chr7 | 21856441 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36560 |
| SBS | Chr7 | 22010689 | C-G | HET | INTRON | |
| SBS | Chr7 | 26268419 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr7 | 6514997 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 7828659 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 13777220 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 19856609 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 7616356 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 16642829 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 3281137 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 3875475 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g07680 |
| SBS | Chr9 | 6889832 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g12190 |
Deletions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 3854189 | 3854190 | 1 | |
| Deletion | Chr7 | 5093824 | 5093825 | 1 | |
| Deletion | Chr1 | 5453365 | 5453368 | 3 | |
| Deletion | Chr10 | 10956592 | 10956595 | 3 | LOC_Os10g21400 |
| Deletion | Chr3 | 13158448 | 13158453 | 5 | |
| Deletion | Chr6 | 21360252 | 21360254 | 2 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 14224874 | 14224876 | 3 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 8533401 | 8545150 | |
| Inversion | Chr4 | 9598154 | 16329056 | LOC_Os04g27610 |
| Inversion | Chr2 | 10297557 | 33235329 | |
| Inversion | Chr4 | 11672235 | 30743435 | |
| Inversion | Chr12 | 13357364 | 13411412 | |
| Inversion | Chr2 | 13634083 | 13704768 | |
| Inversion | Chr1 | 22587319 | 39117896 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 19991105 | Chr4 | 10253749 | 2 |
