Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | W472-5-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download W472-5-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 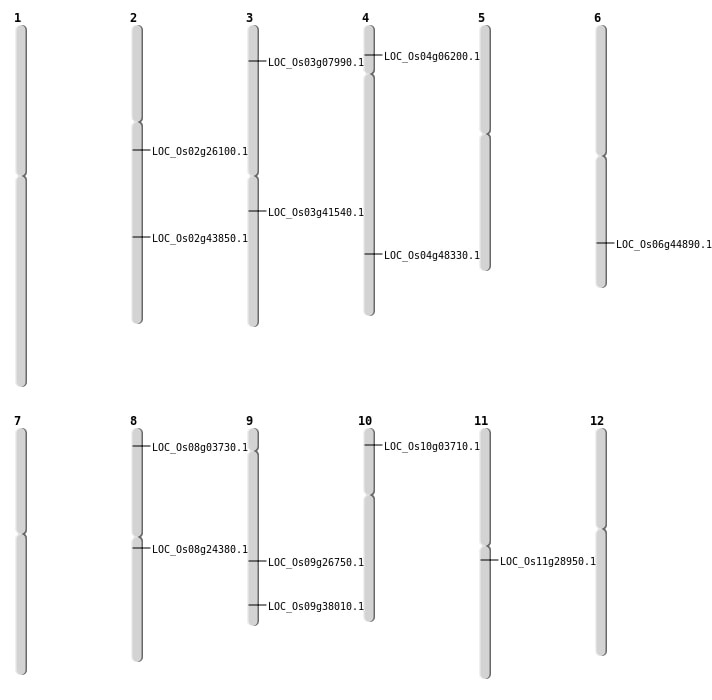
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 30735281 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 403521 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 6344550 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 6583412 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 15910808 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 1674775 | A-G | HOMO | INTRON | |
| SBS | Chr10 | 1676434 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 16753759 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g28950 |
| SBS | Chr12 | 10420240 | C-A | HOMO | INTRON | |
| SBS | Chr12 | 14643541 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 1898014 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 15335829 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g26100 |
| SBS | Chr2 | 26484784 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 28912282 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 29920515 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 13836002 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 16553074 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 19468537 | C-T | HET | INTRON | |
| SBS | Chr3 | 23100797 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g41540 |
| SBS | Chr3 | 35511042 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 4084029 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g07990 |
| SBS | Chr4 | 10934008 | G-A | HET | INTRON | |
| SBS | Chr4 | 15028846 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 16301905 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 16412181 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21523172 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 3222381 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g06200 |
| SBS | Chr4 | 6375813 | T-G | HET | INTRON | |
| SBS | Chr5 | 8133880 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 2617627 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 28489974 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr6 | 4673354 | A-T | HOMO | INTRON | |
| SBS | Chr7 | 18923052 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 27554328 | A-T | HOMO | INTRON | |
| SBS | Chr8 | 27783867 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 10913480 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 20536862 | T-G | HET | INTRON | |
| SBS | Chr9 | 21909834 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g38010 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 562715 | 562723 | 8 | |
| Deletion | Chr5 | 3455060 | 3455063 | 3 | |
| Deletion | Chr6 | 4696963 | 4696964 | 1 | |
| Deletion | Chr7 | 5940519 | 5940592 | 73 | |
| Deletion | Chr3 | 6141620 | 6141641 | 21 | |
| Deletion | Chr1 | 6235578 | 6235580 | 2 | |
| Deletion | Chr9 | 9175711 | 9175726 | 15 | |
| Deletion | Chr8 | 11752416 | 11752418 | 2 | |
| Deletion | Chr2 | 13571953 | 13571967 | 14 | |
| Deletion | Chr10 | 14665197 | 14665209 | 12 | |
| Deletion | Chr8 | 14717097 | 14717130 | 33 | LOC_Os08g24380 |
| Deletion | Chr6 | 14971514 | 14971515 | 1 | |
| Deletion | Chr12 | 21160695 | 21160702 | 7 | |
| Deletion | Chr5 | 22680801 | 22680804 | 3 | |
| Deletion | Chr1 | 24264286 | 24264290 | 4 | |
| Deletion | Chr12 | 25051579 | 25051580 | 1 | |
| Deletion | Chr2 | 26475372 | 26479578 | 4206 | LOC_Os02g43850 |
| Deletion | Chr6 | 27135987 | 27136007 | 20 | LOC_Os06g44890 |
| Deletion | Chr4 | 28814439 | 28814446 | 7 | LOC_Os04g48330 |
| Deletion | Chr7 | 29062534 | 29062545 | 11 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 295499 | 295499 | 1 | |
| Insertion | Chr2 | 4540197 | 4540197 | 1 | |
| Insertion | Chr6 | 1309350 | 1309351 | 2 | |
| Insertion | Chr8 | 3831104 | 3831104 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 1773654 | 1780676 | LOC_Os08g03730 |
| Inversion | Chr4 | 21521469 | 31774983 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1673199 | Chr9 | 16247205 | 2 |
| Translocation | Chr3 | 30629740 | Chr2 | 1615466 |
