Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | W706-3-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download W706-3-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 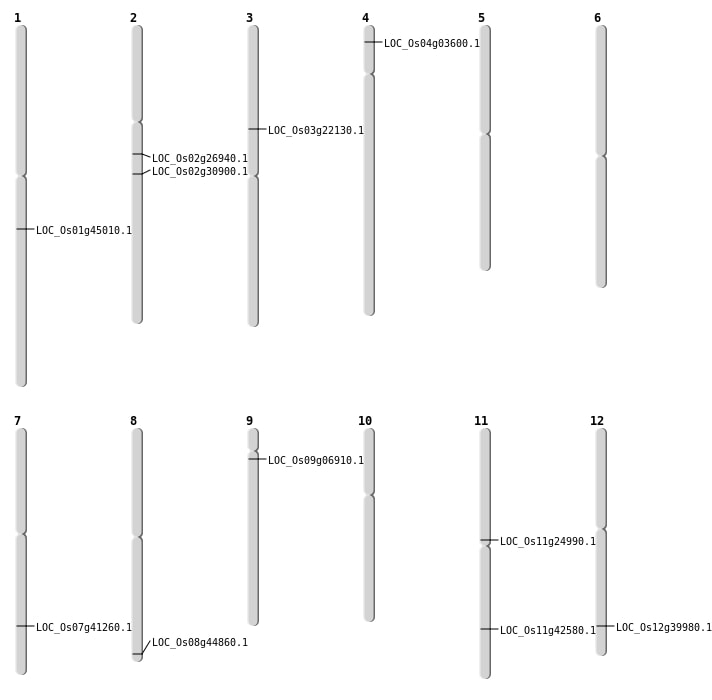
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16513116 | C-A | HET | INTRON | |
| SBS | Chr1 | 19897058 | C-T | HET | INTRON | |
| SBS | Chr1 | 21784575 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 2473638 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 30706488 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 3877972 | C-A | HOMO | INTRON | |
| SBS | Chr10 | 15468601 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 9280906 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 14233305 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g24990 |
| SBS | Chr11 | 6006452 | C-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 12062291 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 13811093 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 23975646 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 24618154 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 24618155 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 24618159 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 15825600 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g26940 |
| SBS | Chr2 | 23658613 | A-T | HOMO | INTRON | |
| SBS | Chr2 | 3241371 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 1574571 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g03600 |
| SBS | Chr4 | 33866523 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 16548188 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 21322554 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 8664270 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 11145573 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 14855469 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 1874048 | G-T | HOMO | INTRON | |
| SBS | Chr7 | 23853253 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 23995263 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 24982855 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 8404042 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 19484300 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 5979375 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 8807498 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 3326572 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g06910 |
| SBS | Chr9 | 5877426 | C-T | HET | INTRON |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2363633 | 2363645 | 12 | |
| Deletion | Chr5 | 2566697 | 2566724 | 27 | |
| Deletion | Chr3 | 2810583 | 2810584 | 1 | |
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr12 | 9374041 | 9374042 | 1 | |
| Deletion | Chr8 | 9887246 | 9887247 | 1 | |
| Deletion | Chr3 | 10377974 | 10377975 | 1 | |
| Deletion | Chr12 | 12210101 | 12210104 | 3 | |
| Deletion | Chr3 | 12687370 | 12687374 | 4 | LOC_Os03g22130 |
| Deletion | Chr11 | 13064956 | 13064967 | 11 | |
| Deletion | Chr9 | 13865291 | 13865298 | 7 | |
| Deletion | Chr1 | 13923211 | 13923216 | 5 | |
| Deletion | Chr12 | 17655879 | 17655880 | 1 | |
| Deletion | Chr2 | 18445004 | 18446095 | 1091 | LOC_Os02g30900 |
| Deletion | Chr10 | 19192690 | 19192693 | 3 | |
| Deletion | Chr12 | 20929797 | 20929812 | 15 | |
| Deletion | Chr7 | 22315276 | 22315303 | 27 | |
| Deletion | Chr8 | 23910215 | 23910218 | 3 | |
| Deletion | Chr11 | 24287834 | 24287842 | 8 | |
| Deletion | Chr12 | 24716432 | 24716435 | 3 | LOC_Os12g39980 |
| Deletion | Chr7 | 24727772 | 24727776 | 4 | LOC_Os07g41260 |
| Deletion | Chr1 | 25538841 | 25538846 | 5 | LOC_Os01g45010 |
| Deletion | Chr11 | 25630828 | 25630833 | 5 | LOC_Os11g42580 |
| Deletion | Chr1 | 29274842 | 29274846 | 4 | |
| Deletion | Chr3 | 30542658 | 30542661 | 3 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11739696 | 11739699 | 4 | |
| Insertion | Chr3 | 5152400 | 5152400 | 1 |
No Inversion
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 2944377 | Chr3 | 24807549 | |
| Translocation | Chr11 | 2945167 | Chr3 | 24807545 | |
| Translocation | Chr8 | 28178265 | Chr2 | 18446079 | 2 |
| Translocation | Chr8 | 28178294 | Chr2 | 18445011 | LOC_Os08g44860 |
