Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-341 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-341 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 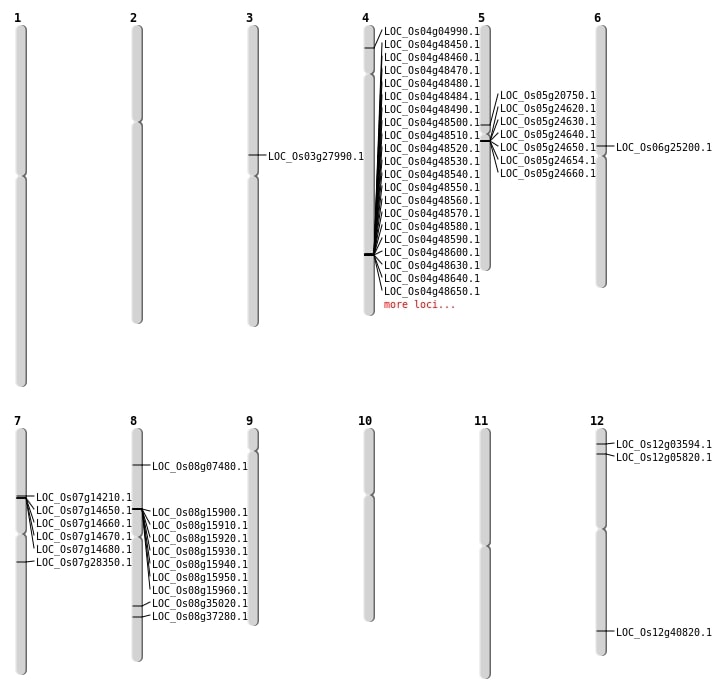
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24954639 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 28906728 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 36107872 | A-T | HET | INTRON | |
| SBS | Chr1 | 38328951 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 38328952 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 20150948 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 23434516 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 25266943 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g40820 |
| SBS | Chr12 | 2683413 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g05820 |
| SBS | Chr2 | 16044003 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 27504531 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 2791140 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 3598467 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 4665209 | T-G | HET | INTRON | |
| SBS | Chr2 | 5221127 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 633025 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 6413292 | T-C | HET | INTRON | |
| SBS | Chr3 | 21897213 | T-C | HET | INTRON | |
| SBS | Chr3 | 2766378 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 20383850 | C-A | HET | INTRON | |
| SBS | Chr4 | 2417906 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g04990 |
| SBS | Chr4 | 33608480 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 5245748 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 12178208 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g20750 |
| SBS | Chr6 | 2617627 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 16549786 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g28350 |
| SBS | Chr7 | 18219417 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 8099578 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g14210 |
| SBS | Chr8 | 23850776 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 13645713 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 13645715 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 18029629 | C-T | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 1437448 | 1437449 | 1 | LOC_Os12g03594 |
| Deletion | Chr3 | 2579652 | 2579660 | 8 | |
| Deletion | Chr9 | 3368176 | 3368178 | 2 | |
| Deletion | Chr8 | 4191675 | 4191692 | 17 | LOC_Os08g07480 |
| Deletion | Chr2 | 5227927 | 5227939 | 12 | |
| Deletion | Chr10 | 8282605 | 8282608 | 3 | |
| Deletion | Chr7 | 8352199 | 8365423 | 13224 | 4 |
| Deletion | Chr10 | 8912169 | 8912172 | 3 | |
| Deletion | Chr8 | 9680001 | 9719000 | 39000 | 7 |
| Deletion | Chr7 | 11959859 | 11959865 | 6 | |
| Deletion | Chr1 | 13159680 | 13159681 | 1 | |
| Deletion | Chr3 | 14090751 | 14090752 | 1 | |
| Deletion | Chr5 | 14216001 | 14264000 | 48000 | 6 |
| Deletion | Chr6 | 14741569 | 14741578 | 9 | LOC_Os06g25200 |
| Deletion | Chr6 | 15512549 | 15512554 | 5 | |
| Deletion | Chr5 | 17346736 | 17346737 | 1 | |
| Deletion | Chr10 | 17416033 | 17416043 | 10 | |
| Deletion | Chr12 | 19457651 | 19457653 | 2 | |
| Deletion | Chr8 | 20974149 | 20974175 | 26 | |
| Deletion | Chr5 | 21658594 | 21659287 | 693 | |
| Deletion | Chr2 | 21782364 | 21782365 | 1 | |
| Deletion | Chr8 | 22060515 | 22060519 | 4 | LOC_Os08g35020 |
| Deletion | Chr10 | 22409273 | 22409274 | 1 | |
| Deletion | Chr8 | 23566037 | 23566039 | 2 | LOC_Os08g37280 |
| Deletion | Chr1 | 24191641 | 24191662 | 21 | |
| Deletion | Chr7 | 27110577 | 27110578 | 1 | |
| Deletion | Chr11 | 27733241 | 27733250 | 9 | |
| Deletion | Chr4 | 28888523 | 29362078 | 473555 | 74 |
| Deletion | Chr4 | 31424484 | 31424489 | 5 | LOC_Os04g52780 |
| Deletion | Chr4 | 31979990 | 31979991 | 1 | LOC_Os04g53660 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 39761084 | 39761084 | 1 | |
| Insertion | Chr3 | 16078768 | 16078769 | 2 | LOC_Os03g27990 |
| Insertion | Chr5 | 20239916 | 20239917 | 2 |
No Inversion
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 22183543 | Chr4 | 7084286 |
