Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-50 [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-50 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 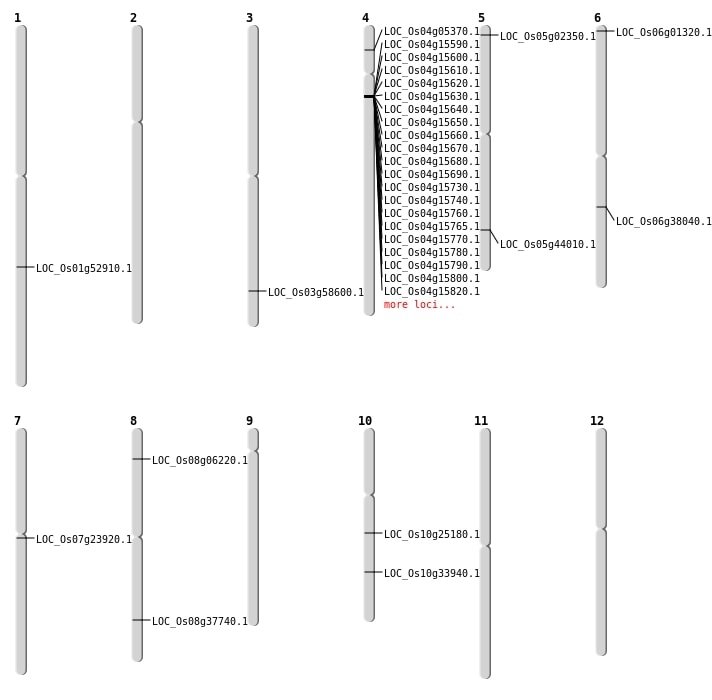
|
| Variant Information |
|---|
Single base substitutions: 10
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19713864 | G-T | Homo | INTERGENIC | |
| SBS | Chr10 | 18056551 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g33940 |
| SBS | Chr11 | 23528183 | A-T | Homo | INTRON | |
| SBS | Chr12 | 22737518 | A-C | Homo | INTERGENIC | |
| SBS | Chr12 | 23826389 | A-T | Homo | INTRON | |
| SBS | Chr3 | 1479076 | T-C | Homo | INTRON | |
| SBS | Chr4 | 8326043 | G-A | Homo | INTERGENIC | |
| SBS | Chr5 | 21203685 | T-C | Homo | INTRON | |
| SBS | Chr8 | 8159977 | T-G | Homo | UTR_3_PRIME | |
| SBS | Chr9 | 18007372 | C-T | Homo | INTRON |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1760273 | 1760278 | 5 | |
| Deletion | Chr10 | 1910221 | 1910224 | 3 | |
| Deletion | Chr4 | 2677170 | 2677178 | 8 | LOC_Os04g05370 |
| Deletion | Chr8 | 3431722 | 3431723 | 1 | LOC_Os08g06220 |
| Deletion | Chr4 | 8477001 | 8863000 | 386000 | 64 |
| Deletion | Chr7 | 9287772 | 9287805 | 33 | |
| Deletion | Chr2 | 11631001 | 11643000 | 12000 | |
| Deletion | Chr10 | 12999606 | 12999619 | 13 | LOC_Os10g25180 |
| Deletion | Chr7 | 13520009 | 13520014 | 5 | LOC_Os07g23920 |
| Deletion | Chr6 | 13956518 | 13956526 | 8 | |
| Deletion | Chr9 | 14446446 | 14446447 | 1 | |
| Deletion | Chr6 | 15135009 | 15135011 | 2 | |
| Deletion | Chr2 | 16017874 | 16017876 | 2 | |
| Deletion | Chr11 | 18769313 | 18769317 | 4 | |
| Deletion | Chr3 | 22481238 | 22481241 | 3 | |
| Deletion | Chr6 | 22492056 | 22492072 | 16 | LOC_Os06g38040 |
| Deletion | Chr8 | 23904092 | 23904094 | 2 | LOC_Os08g37740 |
| Deletion | Chr12 | 25227788 | 25227791 | 3 | |
| Deletion | Chr12 | 27478963 | 27478964 | 1 | |
| Deletion | Chr6 | 27897723 | 27897727 | 4 | |
| Deletion | Chr1 | 30422285 | 30422289 | 4 | LOC_Os01g52910 |
| Deletion | Chr1 | 31341027 | 31341028 | 1 | |
| Deletion | Chr3 | 32795544 | 32795565 | 21 | |
| Deletion | Chr3 | 33375023 | 33375024 | 1 | LOC_Os03g58600 |
Insertions: 36
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10172192 | 10172193 | 2 | |
| Insertion | Chr1 | 23912303 | 23912304 | 2 | |
| Insertion | Chr1 | 2549378 | 2549399 | 22 | |
| Insertion | Chr10 | 13690346 | 13690347 | 2 | |
| Insertion | Chr11 | 16367368 | 16367369 | 2 | |
| Insertion | Chr11 | 18804838 | 18804841 | 4 | |
| Insertion | Chr11 | 25885371 | 25885374 | 4 | |
| Insertion | Chr11 | 26038899 | 26038900 | 2 | |
| Insertion | Chr11 | 26129615 | 26129616 | 2 | |
| Insertion | Chr11 | 7038123 | 7038125 | 3 | |
| Insertion | Chr12 | 23461991 | 23461992 | 2 | |
| Insertion | Chr2 | 16958987 | 16958989 | 3 | |
| Insertion | Chr2 | 17850754 | 17850755 | 2 | |
| Insertion | Chr2 | 6394878 | 6394879 | 2 | |
| Insertion | Chr3 | 10202816 | 10202821 | 6 | |
| Insertion | Chr3 | 14193059 | 14193099 | 41 | |
| Insertion | Chr3 | 24268154 | 24268175 | 22 | |
| Insertion | Chr3 | 26901345 | 26901345 | 1 | |
| Insertion | Chr3 | 381152 | 381157 | 6 | |
| Insertion | Chr4 | 17788550 | 17788552 | 3 | |
| Insertion | Chr4 | 20356376 | 20356377 | 2 | |
| Insertion | Chr4 | 20701596 | 20701597 | 2 | |
| Insertion | Chr4 | 21426409 | 21426416 | 8 | |
| Insertion | Chr4 | 347730 | 347733 | 4 | |
| Insertion | Chr5 | 16387468 | 16387468 | 1 | |
| Insertion | Chr5 | 267129 | 267129 | 1 | |
| Insertion | Chr5 | 763686 | 763687 | 2 | LOC_Os05g02350 |
| Insertion | Chr5 | 7814999 | 7815000 | 2 | |
| Insertion | Chr6 | 4464678 | 4464681 | 4 | |
| Insertion | Chr7 | 17135544 | 17135544 | 1 | |
| Insertion | Chr7 | 3643477 | 3643479 | 3 | |
| Insertion | Chr7 | 8045906 | 8045909 | 4 | |
| Insertion | Chr8 | 20425353 | 20425354 | 2 | |
| Insertion | Chr8 | 970783 | 970788 | 6 | |
| Insertion | Chr9 | 10786503 | 10786506 | 4 | |
| Insertion | Chr9 | 17152786 | 17152813 | 28 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 11640727 | 13153786 | |
| Inversion | Chr2 | 12681053 | 13151441 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 200212 | Chr5 | 25603983 | 2 |
| Translocation | Chr6 | 205188 | Chr5 | 25603982 | 2 |
