Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1124-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1124-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 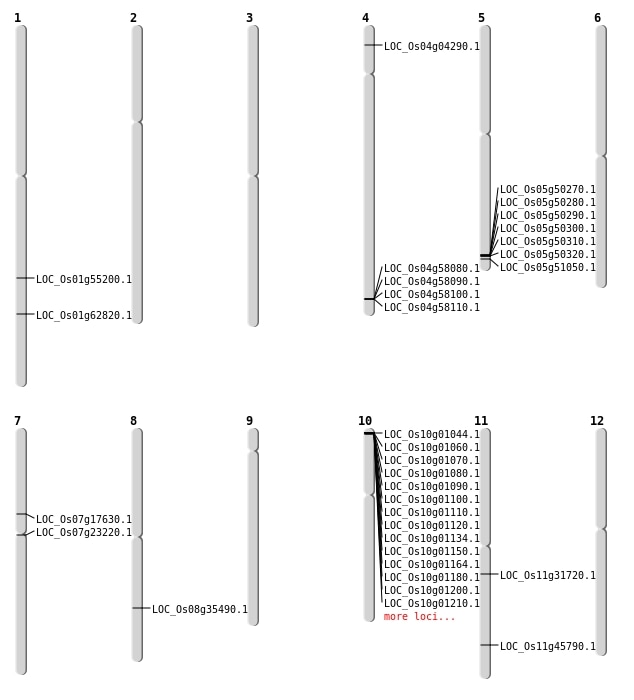
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13308981 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 17043865 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 20225304 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 3493027 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 36386565 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g62820 |
| SBS | Chr1 | 6280259 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr11 | 10691514 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 11101912 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 18597147 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g31720 |
| SBS | Chr11 | 20289955 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 27705912 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g45790 |
| SBS | Chr11 | 5165402 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 12562005 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 1583162 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 1835714 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 1565670 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 16497120 | G-A | HET | INTRON | |
| SBS | Chr2 | 5177504 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 8620687 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 18668882 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 9934563 | C-T | HET | INTRON | |
| SBS | Chr4 | 2007049 | C-A | HOMO | STOP_GAINED | LOC_Os04g04290 |
| SBS | Chr4 | 8089257 | T-A | HOMO | UTR_5_PRIME | |
| SBS | Chr5 | 16527997 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 27932748 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 6394367 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 8491972 | T-A | HET | INTRON | |
| SBS | Chr7 | 10416068 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g17630 |
| SBS | Chr8 | 161789 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 188810 | T-A | HOMO | INTRON | |
| SBS | Chr8 | 19837162 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 22362782 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g35490 |
| SBS | Chr8 | 4102973 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 8314448 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 9295657 | T-A | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 55001 | 325000 | 270000 | 40 |
| Deletion | Chr3 | 4937660 | 4937671 | 11 | |
| Deletion | Chr10 | 5990645 | 5990647 | 2 | |
| Deletion | Chr5 | 10207440 | 10207450 | 10 | |
| Deletion | Chr9 | 10828356 | 10828463 | 107 | |
| Deletion | Chr3 | 11249633 | 11249646 | 13 | |
| Deletion | Chr7 | 13088211 | 13088212 | 1 | LOC_Os07g23220 |
| Deletion | Chr11 | 13091414 | 13091415 | 1 | |
| Deletion | Chr1 | 17676109 | 17676112 | 3 | |
| Deletion | Chr2 | 19577666 | 19577669 | 3 | |
| Deletion | Chr6 | 21958825 | 21958831 | 6 | |
| Deletion | Chr8 | 22002998 | 22003015 | 17 | |
| Deletion | Chr5 | 22439261 | 22439271 | 10 | |
| Deletion | Chr11 | 23110694 | 23110703 | 9 | |
| Deletion | Chr6 | 24599495 | 24599507 | 12 | |
| Deletion | Chr1 | 25113728 | 25113730 | 2 | |
| Deletion | Chr2 | 27945741 | 27945764 | 23 | |
| Deletion | Chr5 | 28810461 | 28837949 | 27488 | 6 |
| Deletion | Chr4 | 29365741 | 29365747 | 6 | |
| Deletion | Chr4 | 34590915 | 34611994 | 21079 | 4 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 1551778 | 1551778 | 1 | |
| Insertion | Chr2 | 24894725 | 24894727 | 3 | |
| Insertion | Chr7 | 7499018 | 7499026 | 9 | |
| Insertion | Chr9 | 15545377 | 15545378 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 7301144 | 7397105 | |
| Inversion | Chr1 | 7301169 | 7397110 | |
| Inversion | Chr5 | 8159181 | 29294247 | LOC_Os05g51050 |
| Inversion | Chr5 | 8159841 | 29294234 | LOC_Os05g51050 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 2121768 | Chr1 | 31762827 | LOC_Os01g55200 |
| Translocation | Chr5 | 2122717 | Chr1 | 31762794 | LOC_Os01g55200 |
