Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1135-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1135-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 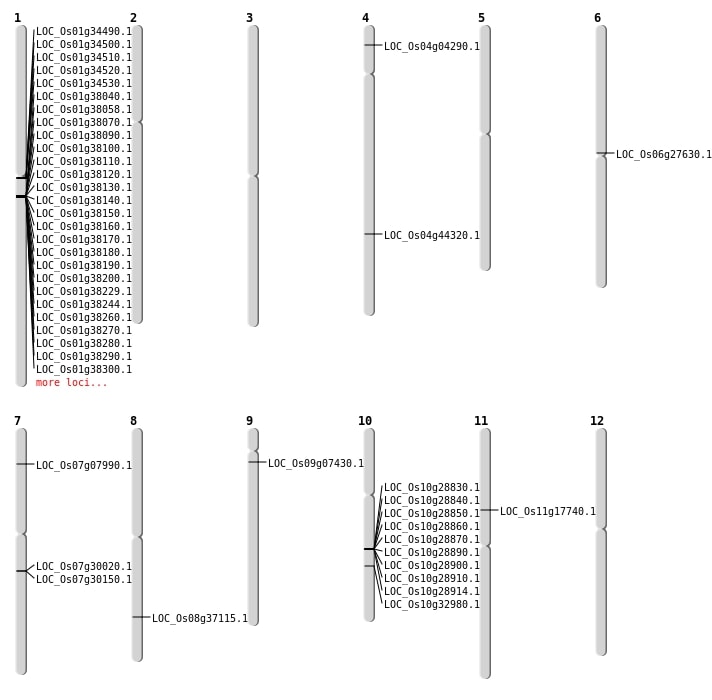
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10954371 | G-A | HET | INTRON | |
| SBS | Chr1 | 17464025 | C-A | HET | INTRON | |
| SBS | Chr1 | 38558000 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 38558002 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g66379 |
| SBS | Chr10 | 17265405 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g32980 |
| SBS | Chr10 | 17883302 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 6778900 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 13799841 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 22630088 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 23816458 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 9981774 | G-A | HET | INTRON | |
| SBS | Chr12 | 3993644 | C-T | HET | INTRON | |
| SBS | Chr2 | 30653835 | C-A | HOMO | INTRON | |
| SBS | Chr2 | 31455036 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 3676302 | C-T | HET | INTRON | |
| SBS | Chr4 | 2009016 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g04290 |
| SBS | Chr5 | 6417555 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 16719969 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 16719970 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 21930601 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 7837106 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 19681060 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 11657418 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 11972150 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 8233485 | C-T | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 1779535 | 1779569 | 34 | |
| Deletion | Chr7 | 2508686 | 2508697 | 11 | |
| Deletion | Chr7 | 4030117 | 4030120 | 3 | LOC_Os07g07990 |
| Deletion | Chr11 | 5673100 | 5673111 | 11 | |
| Deletion | Chr12 | 8459840 | 8459842 | 2 | |
| Deletion | Chr11 | 9917671 | 9917673 | 2 | LOC_Os11g17740 |
| Deletion | Chr6 | 12813952 | 12813972 | 20 | |
| Deletion | Chr10 | 15027001 | 15072000 | 45000 | 9 |
| Deletion | Chr6 | 15630732 | 15632324 | 1592 | LOC_Os06g27630 |
| Deletion | Chr10 | 16090738 | 16090739 | 1 | |
| Deletion | Chr11 | 17308907 | 17308908 | 1 | |
| Deletion | Chr3 | 17541140 | 17541141 | 1 | |
| Deletion | Chr1 | 18185617 | 18185619 | 2 | |
| Deletion | Chr1 | 19016296 | 19044907 | 28611 | 5 |
| Deletion | Chr7 | 19026030 | 19026032 | 2 | |
| Deletion | Chr2 | 19858325 | 19858326 | 1 | |
| Deletion | Chr1 | 20022648 | 20022650 | 2 | |
| Deletion | Chr8 | 20836443 | 20836445 | 2 | |
| Deletion | Chr1 | 21314001 | 21825000 | 511000 | 76 |
| Deletion | Chr8 | 23451578 | 23451587 | 9 | LOC_Os08g37115 |
| Deletion | Chr4 | 24132276 | 24132289 | 13 | |
| Deletion | Chr7 | 25370686 | 25370715 | 29 | |
| Deletion | Chr3 | 25580816 | 25580829 | 13 | |
| Deletion | Chr3 | 26689070 | 26689078 | 8 | |
| Deletion | Chr6 | 27010691 | 27010697 | 6 | |
| Deletion | Chr4 | 27753065 | 27753066 | 1 | |
| Deletion | Chr4 | 28962794 | 28962795 | 1 | |
| Deletion | Chr1 | 29365632 | 29365637 | 5 | LOC_Os01g51120 |
| Deletion | Chr2 | 33673050 | 33673081 | 31 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 23050004 | 23050005 | 2 | |
| Insertion | Chr12 | 20273258 | 20273259 | 2 | |
| Insertion | Chr12 | 23551114 | 23551115 | 2 | |
| Insertion | Chr2 | 12353499 | 12353502 | 4 | |
| Insertion | Chr4 | 18959179 | 18959180 | 2 | |
| Insertion | Chr4 | 26251100 | 26251100 | 1 | LOC_Os04g44320 |
| Insertion | Chr6 | 27095370 | 27095370 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 15030310 | 16265459 | |
| Inversion | Chr7 | 16388242 | 17687708 | LOC_Os07g30020 |
| Inversion | Chr7 | 16388401 | 17687730 | LOC_Os07g30020 |
| Inversion | Chr1 | 37890259 | 38156305 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 3712680 | Chr1 | 21825815 | 2 |
| Translocation | Chr4 | 6944309 | Chr1 | 37473759 | |
| Translocation | Chr4 | 6944318 | Chr1 | 37472724 | LOC_Os01g64590 |
| Translocation | Chr7 | 16718148 | Chr1 | 21760272 | LOC_Os01g38780 |
| Translocation | Chr7 | 17792809 | Chr1 | 21760573 | 2 |
