Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1228-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1228-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 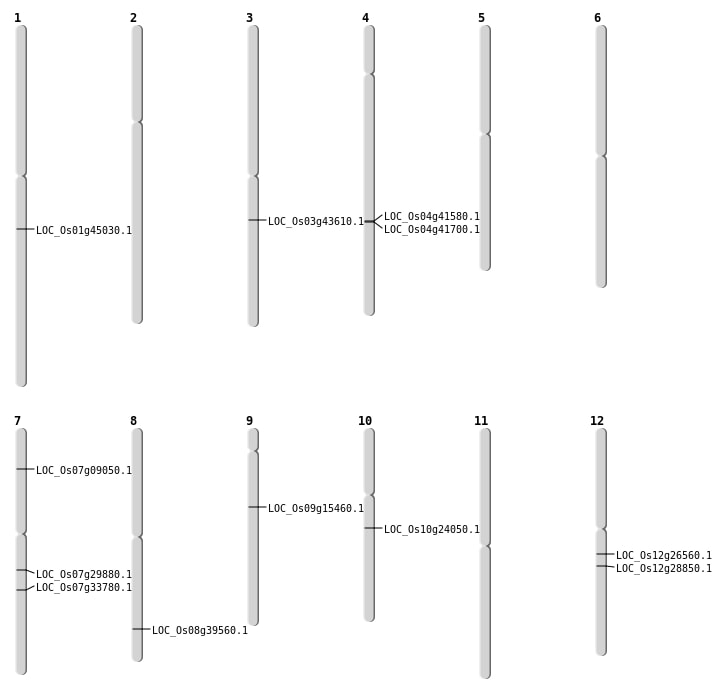
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12269615 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 16467630 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 19504665 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 25555339 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g45030 |
| SBS | Chr10 | 12321439 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24050 |
| SBS | Chr10 | 18991902 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 12234861 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14422711 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 15527916 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26560 |
| SBS | Chr12 | 7417691 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 12496415 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 19813461 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 4948860 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 5540414 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 15142298 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 24374368 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g43610 |
| SBS | Chr3 | 29385404 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 29385413 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 34617107 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 35510681 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 6506541 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 10513146 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 34904945 | A-T | HET | INTRON | |
| SBS | Chr4 | 4335314 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 7371255 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 14171272 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 21656615 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 25937118 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr5 | 29053641 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 8664270 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 9515308 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 23862373 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 5820501 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 17293881 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 18746220 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 22657806 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 23995263 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 24982855 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 6940085 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5366721 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 9307587 | G-T | HOMO | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 190241 | 194015 | 3774 | |
| Deletion | Chr1 | 981531 | 981532 | 1 | |
| Deletion | Chr3 | 1677588 | 1677599 | 11 | |
| Deletion | Chr3 | 3568420 | 3568423 | 3 | |
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr7 | 4701743 | 4701767 | 24 | |
| Deletion | Chr1 | 5745430 | 5745431 | 1 | |
| Deletion | Chr11 | 6023213 | 6023225 | 12 | |
| Deletion | Chr11 | 6196366 | 6196385 | 19 | |
| Deletion | Chr11 | 6198150 | 6198462 | 312 | |
| Deletion | Chr9 | 9460332 | 9460335 | 3 | LOC_Os09g15460 |
| Deletion | Chr9 | 10498201 | 10498210 | 9 | |
| Deletion | Chr1 | 10951576 | 10951602 | 26 | |
| Deletion | Chr1 | 11547217 | 11547222 | 5 | |
| Deletion | Chr2 | 12079171 | 12079172 | 1 | |
| Deletion | Chr11 | 14036819 | 14036849 | 30 | |
| Deletion | Chr1 | 17049254 | 17049255 | 1 | |
| Deletion | Chr11 | 17351757 | 17351758 | 1 | |
| Deletion | Chr10 | 17531854 | 17531873 | 19 | |
| Deletion | Chr7 | 18750503 | 18750505 | 2 | |
| Deletion | Chr7 | 19282875 | 19282881 | 6 | |
| Deletion | Chr7 | 20217898 | 20217901 | 3 | LOC_Os07g33780 |
| Deletion | Chr2 | 20692348 | 20692357 | 9 | |
| Deletion | Chr9 | 21141851 | 21141853 | 2 | |
| Deletion | Chr6 | 23171396 | 23171429 | 33 | |
| Deletion | Chr6 | 23862817 | 23862820 | 3 | |
| Deletion | Chr8 | 25024480 | 25024484 | 4 | LOC_Os08g39560 |
| Deletion | Chr1 | 25177474 | 25177483 | 9 | |
| Deletion | Chr2 | 26676160 | 26676161 | 1 | |
| Deletion | Chr4 | 29051598 | 29051602 | 4 |
Insertions: 5
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 4729755 | 5772195 | LOC_Os07g09050 |
| Inversion | Chr8 | 16435701 | 17326152 | |
| Inversion | Chr12 | 17054932 | 17771111 | LOC_Os12g28850 |
| Inversion | Chr8 | 17333232 | 18002830 | |
| Inversion | Chr7 | 17587815 | 17740088 | LOC_Os07g29880 |
| Inversion | Chr4 | 24669500 | 24721362 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 18628274 | Chr5 | 9638214 |
