Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1229-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1229-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 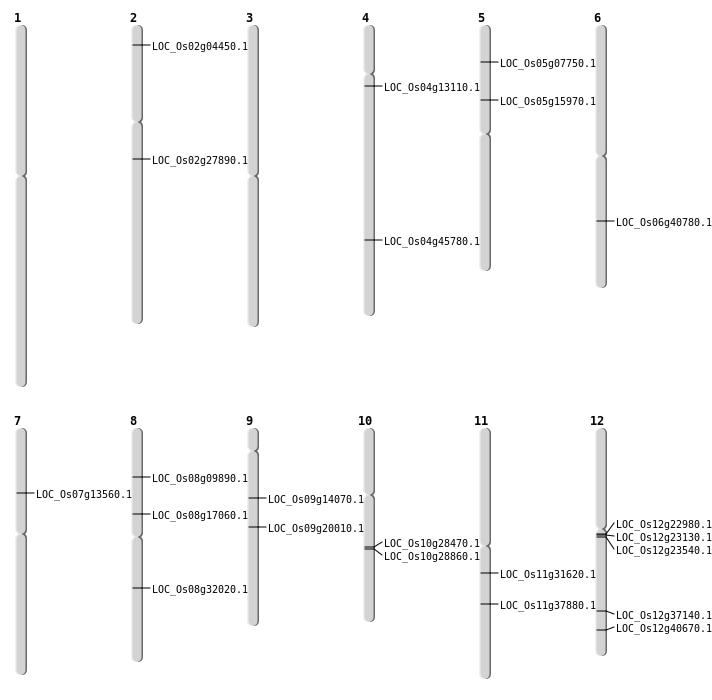
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 40699469 | C-T | HOMO | INTRON | |
| SBS | Chr10 | 10009259 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11580977 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 14303960 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 13083676 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23130 |
| SBS | Chr12 | 17178263 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 22502655 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 22502658 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 22765441 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g37140 |
| SBS | Chr12 | 25180483 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g40670 |
| SBS | Chr12 | 26465567 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 26465568 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 4618841 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 5448730 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 19994897 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 3614281 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 24300436 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 27094906 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g45780 |
| SBS | Chr4 | 35498128 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 5033893 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 6285600 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 15390458 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 9014962 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15970 |
| SBS | Chr6 | 24316002 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g40780 |
| SBS | Chr7 | 12685468 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 13489243 | T-C | HET | INTRON | |
| SBS | Chr7 | 25878110 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 28706966 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 19863755 | C-T | HET | STOP_GAINED | LOC_Os08g32020 |
| SBS | Chr8 | 21337303 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 11395074 | A-T | HOMO | UTR_3_PRIME | |
| SBS | Chr9 | 11414275 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 3172181 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 826549 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 8330241 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g14070 |
| SBS | Chr9 | 900638 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 9188696 | A-C | HET | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1971014 | 1972409 | 1395 | LOC_Os02g04450 |
| Deletion | Chr7 | 2938249 | 2938250 | 1 | |
| Deletion | Chr5 | 4178773 | 4178775 | 2 | LOC_Os05g07750 |
| Deletion | Chr8 | 5708668 | 5708671 | 3 | LOC_Os08g09890 |
| Deletion | Chr4 | 7233531 | 7233541 | 10 | LOC_Os04g13110 |
| Deletion | Chr7 | 7773468 | 7773473 | 5 | LOC_Os07g13560 |
| Deletion | Chr5 | 8247562 | 8247563 | 1 | |
| Deletion | Chr1 | 10098199 | 10098202 | 3 | |
| Deletion | Chr8 | 10296066 | 10296067 | 1 | |
| Deletion | Chr8 | 10444460 | 10444461 | 1 | LOC_Os08g17060 |
| Deletion | Chr1 | 11747225 | 11747226 | 1 | |
| Deletion | Chr9 | 11988770 | 11988776 | 6 | LOC_Os09g20010 |
| Deletion | Chr11 | 12899369 | 12899380 | 11 | |
| Deletion | Chr12 | 12991680 | 12991744 | 64 | LOC_Os12g22980 |
| Deletion | Chr12 | 13326346 | 13326347 | 1 | LOC_Os12g23540 |
| Deletion | Chr10 | 14812865 | 14812867 | 2 | LOC_Os10g28470 |
| Deletion | Chr6 | 16506111 | 16506112 | 1 | |
| Deletion | Chr11 | 18487015 | 18487030 | 15 | LOC_Os11g31620 |
| Deletion | Chr11 | 22454602 | 22454606 | 4 | LOC_Os11g37880 |
| Deletion | Chr8 | 23138860 | 23138865 | 5 | |
| Deletion | Chr4 | 30433565 | 30433569 | 4 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 31489786 | 31489786 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 12472954 | 15045219 | LOC_Os10g28860 |
| Inversion | Chr2 | 16075710 | 16509962 | LOC_Os02g27890 |
| Inversion | Chr2 | 16075723 | 16509977 | LOC_Os02g27890 |
No Translocation
