Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1258-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1258-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 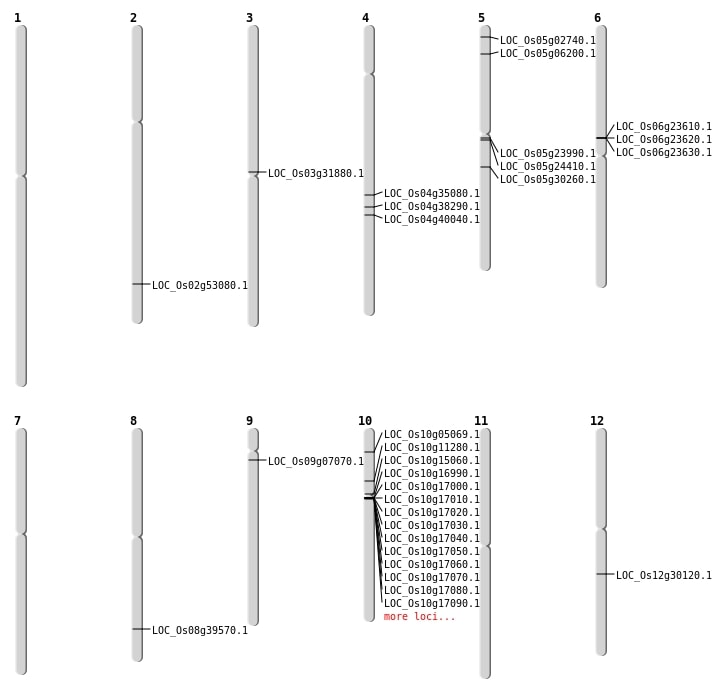
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 36853369 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 4580688 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 8701008 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11927853 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 7883421 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g15060 |
| SBS | Chr11 | 25229965 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 2617955 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 18045129 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g30120 |
| SBS | Chr2 | 24662479 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 9109901 | C-A | HET | INTRON | |
| SBS | Chr3 | 18239542 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g31880 |
| SBS | Chr3 | 2205421 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 33029503 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 3849135 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 14155898 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 21324818 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 21324822 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g35080 |
| SBS | Chr4 | 2155663 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 9081809 | G-C | HOMO | INTRON | |
| SBS | Chr5 | 14967461 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 22965347 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 26462003 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 4082372 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 8720311 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 8256701 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 18151365 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 24152418 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 24708051 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 406748 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 3427107 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07070 |
| SBS | Chr9 | 3427108 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 7897886 | A-G | HOMO | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 702768 | 702773 | 5 | |
| Deletion | Chr12 | 1235460 | 1235484 | 24 | |
| Deletion | Chr5 | 3122092 | 3122105 | 13 | LOC_Os05g06200 |
| Deletion | Chr11 | 3434537 | 3434539 | 2 | |
| Deletion | Chr4 | 3703161 | 3703163 | 2 | |
| Deletion | Chr11 | 3828535 | 3828544 | 9 | |
| Deletion | Chr3 | 3848901 | 3848912 | 11 | |
| Deletion | Chr4 | 5107588 | 5107589 | 1 | |
| Deletion | Chr8 | 5586401 | 5586402 | 1 | |
| Deletion | Chr10 | 6259016 | 6259020 | 4 | LOC_Os10g11280 |
| Deletion | Chr12 | 6810152 | 6810160 | 8 | |
| Deletion | Chr10 | 8506001 | 8664000 | 158000 | 21 |
| Deletion | Chr12 | 12465494 | 12465516 | 22 | |
| Deletion | Chr6 | 13772548 | 13777419 | 4871 | 3 |
| Deletion | Chr9 | 14001163 | 14001187 | 24 | |
| Deletion | Chr3 | 14495324 | 14495326 | 2 | |
| Deletion | Chr7 | 14716916 | 14716927 | 11 | |
| Deletion | Chr5 | 17538135 | 17538142 | 7 | LOC_Os05g30260 |
| Deletion | Chr11 | 18092514 | 18092531 | 17 | |
| Deletion | Chr4 | 18971011 | 18971035 | 24 | |
| Deletion | Chr2 | 19617462 | 19617472 | 10 | |
| Deletion | Chr4 | 22785153 | 22785162 | 9 | LOC_Os04g38290 |
| Deletion | Chr4 | 23829802 | 23829803 | 1 | LOC_Os04g40040 |
| Deletion | Chr8 | 25035215 | 25035216 | 1 | LOC_Os08g39570 |
| Deletion | Chr2 | 29362956 | 29362959 | 3 | |
| Deletion | Chr2 | 35623982 | 35624001 | 19 | |
| Deletion | Chr1 | 41648714 | 41648722 | 8 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 13040938 | 13040944 | 7 | |
| Insertion | Chr4 | 21130579 | 21130580 | 2 | |
| Insertion | Chr8 | 16438848 | 16438855 | 8 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 2429077 | 2483020 | LOC_Os10g05069 |
| Inversion | Chr5 | 12316646 | 14105156 | LOC_Os05g24410 |
| Inversion | Chr5 | 13819941 | 14105142 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 986725 | Chr2 | 32475676 | 2 |
| Translocation | Chr12 | 15304618 | Chr4 | 1265206 | |
| Translocation | Chr11 | 19390024 | Chr3 | 4264017 | |
| Translocation | Chr5 | 22550101 | Chr3 | 23592967 |
