Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1262-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1262-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 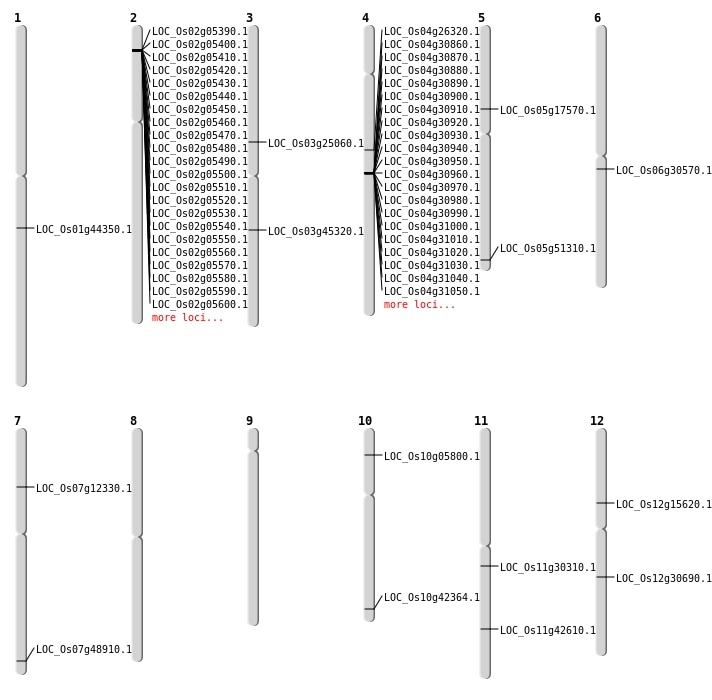
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 4550369 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 4747817 | A-G | HET | INTRON | |
| SBS | Chr10 | 22806021 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g42364 |
| SBS | Chr12 | 14693223 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 14693241 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 15706585 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 18432680 | A-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g30690 |
| SBS | Chr12 | 27162429 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 11313534 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 21845783 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 6697909 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 25588216 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g45320 |
| SBS | Chr3 | 29419488 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 16221189 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 16359426 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 25834734 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 29641694 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 10091870 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17570 |
| SBS | Chr5 | 4054398 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 13833746 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 17689497 | G-T | HET | STOP_GAINED | LOC_Os06g30570 |
| SBS | Chr6 | 5434705 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 8161206 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 8875050 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 9637563 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 15378907 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 15378911 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 831442 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 22969480 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 6738443 | G-A | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1521977 | 1521985 | 8 | |
| Deletion | Chr9 | 2405570 | 2405575 | 5 | |
| Deletion | Chr2 | 2592001 | 3175000 | 583000 | 98 |
| Deletion | Chr12 | 3079312 | 3079335 | 23 | |
| Deletion | Chr7 | 3264558 | 3264564 | 6 | |
| Deletion | Chr1 | 4669809 | 4669811 | 2 | |
| Deletion | Chr11 | 5851223 | 5851233 | 10 | |
| Deletion | Chr2 | 6835836 | 6835837 | 1 | |
| Deletion | Chr2 | 7720789 | 7720790 | 1 | LOC_Os02g14130 |
| Deletion | Chr12 | 8921079 | 8921084 | 5 | LOC_Os12g15620 |
| Deletion | Chr10 | 12041119 | 12041124 | 5 | |
| Deletion | Chr3 | 14302760 | 14302763 | 3 | LOC_Os03g25060 |
| Deletion | Chr4 | 15342785 | 15342786 | 1 | LOC_Os04g26320 |
| Deletion | Chr7 | 15360970 | 15361008 | 38 | |
| Deletion | Chr11 | 17601395 | 17601399 | 4 | LOC_Os11g30310 |
| Deletion | Chr7 | 17776043 | 17776047 | 4 | |
| Deletion | Chr1 | 18068605 | 18068617 | 12 | |
| Deletion | Chr4 | 18433001 | 18580000 | 147000 | 22 |
| Deletion | Chr4 | 18844341 | 18844357 | 16 | |
| Deletion | Chr6 | 19818562 | 19818565 | 3 | |
| Deletion | Chr2 | 21814741 | 21814743 | 2 | |
| Deletion | Chr8 | 22551778 | 22551797 | 19 | |
| Deletion | Chr8 | 24397792 | 24397801 | 9 | |
| Deletion | Chr8 | 27365830 | 27365907 | 77 | |
| Deletion | Chr2 | 28071515 | 28071516 | 1 | |
| Deletion | Chr7 | 29269737 | 29269738 | 1 | LOC_Os07g48910 |
| Deletion | Chr5 | 29434700 | 29434706 | 6 | LOC_Os05g51310 |
| Deletion | Chr3 | 36259793 | 36259801 | 8 | |
| Deletion | Chr1 | 39604030 | 39604034 | 4 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 3680394 | 3680394 | 1 | |
| Insertion | Chr4 | 20440159 | 20440159 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 25445817 | 26307622 | LOC_Os01g44350 |
| Inversion | Chr1 | 25445836 | 26307632 | LOC_Os01g44350 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2933877 | Chr7 | 6953401 | 2 |
| Translocation | Chr10 | 2933889 | Chr7 | 6955434 | 2 |
| Translocation | Chr9 | 8941257 | Chr1 | 30237197 | |
| Translocation | Chr12 | 15284223 | Chr10 | 15776444 | |
| Translocation | Chr11 | 25650477 | Chr7 | 9569138 | LOC_Os11g42610 |
