Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1272-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1272-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 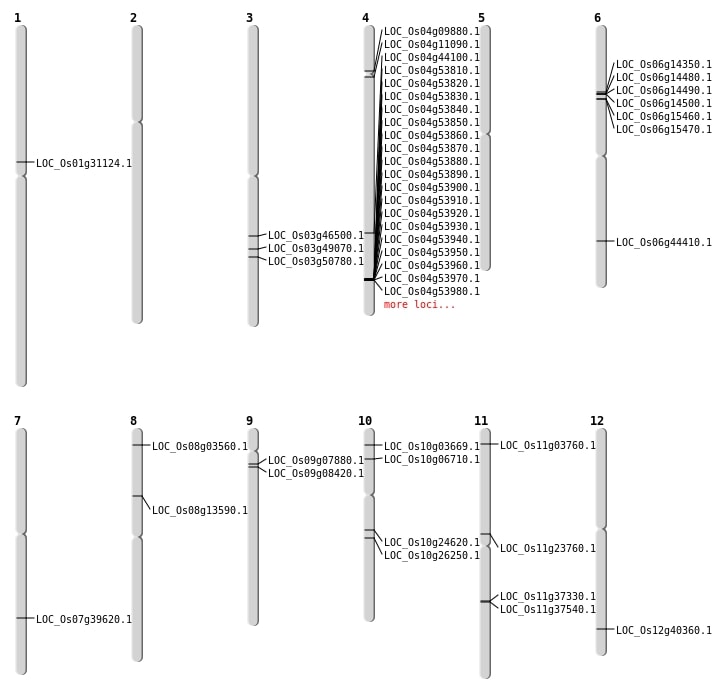
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11344916 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 25247238 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 26876338 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 27666127 | C-T | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr1 | 8210443 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 1643330 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g03669 |
| SBS | Chr11 | 1247202 | G-A | HET | INTRON | |
| SBS | Chr11 | 13437945 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g23760 |
| SBS | Chr11 | 1481170 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 7444305 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 9680358 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 128555 | C-T | HET | INTRON | |
| SBS | Chr2 | 13937360 | T-C | HET | INTRON | |
| SBS | Chr3 | 18712750 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 27956670 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g49070 |
| SBS | Chr3 | 28315755 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 10384185 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 27604008 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 33710198 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 5312729 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g09880 |
| SBS | Chr4 | 6034714 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g11090 |
| SBS | Chr5 | 26890059 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 10822071 | G-A | HET | INTRON | |
| SBS | Chr6 | 13952718 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 26810934 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g44410 |
| SBS | Chr6 | 30436203 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 8616303 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 10222575 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 13497752 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 20783800 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 23100994 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 27294551 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 7828659 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 13777220 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 17997157 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 4383873 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g08420 |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1656665 | 1656669 | 4 | LOC_Os08g03560 |
| Deletion | Chr5 | 2184407 | 2184415 | 8 | |
| Deletion | Chr6 | 3020073 | 3020077 | 4 | |
| Deletion | Chr10 | 3473742 | 3473743 | 1 | LOC_Os10g06710 |
| Deletion | Chr9 | 4005437 | 4005442 | 5 | LOC_Os09g07880 |
| Deletion | Chr1 | 4490809 | 4490816 | 7 | |
| Deletion | Chr10 | 6703058 | 6703066 | 8 | |
| Deletion | Chr7 | 7051152 | 7051153 | 1 | |
| Deletion | Chr8 | 8079036 | 8079037 | 1 | LOC_Os08g13590 |
| Deletion | Chr6 | 8141001 | 8150000 | 9000 | 3 |
| Deletion | Chr9 | 8819107 | 8819108 | 1 | |
| Deletion | Chr10 | 12636604 | 12636838 | 234 | LOC_Os10g24620 |
| Deletion | Chr5 | 13287153 | 13287156 | 3 | |
| Deletion | Chr10 | 13593503 | 13593515 | 12 | LOC_Os10g26250 |
| Deletion | Chr5 | 15319815 | 15319817 | 2 | |
| Deletion | Chr7 | 15450963 | 15450964 | 1 | |
| Deletion | Chr1 | 17025131 | 17025133 | 2 | LOC_Os01g31124 |
| Deletion | Chr11 | 22041003 | 22041011 | 8 | LOC_Os11g37330 |
| Deletion | Chr1 | 22968597 | 22968605 | 8 | |
| Deletion | Chr7 | 23009928 | 23009992 | 64 | |
| Deletion | Chr4 | 26111836 | 26111839 | 3 | LOC_Os04g44100 |
| Deletion | Chr3 | 26314526 | 26314529 | 3 | LOC_Os03g46500 |
| Deletion | Chr1 | 28269005 | 28269015 | 10 | |
| Deletion | Chr3 | 28990565 | 28990571 | 6 | LOC_Os03g50780 |
| Deletion | Chr4 | 30736275 | 30736391 | 116 | |
| Deletion | Chr4 | 32063159 | 32175544 | 112385 | 20 |
| Deletion | Chr4 | 32223758 | 32248306 | 24548 | 6 |
| Deletion | Chr4 | 32763001 | 32807000 | 44000 | 9 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 24994311 | 24994312 | 2 | LOC_Os12g40360 |
| Insertion | Chr3 | 11892663 | 11892663 | 1 | |
| Insertion | Chr4 | 31490833 | 31490835 | 3 | |
| Insertion | Chr9 | 4781773 | 4781774 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 8003720 | 8140678 | LOC_Os06g14350 |
| Inversion | Chr6 | 8149686 | 8777209 | LOC_Os06g15460 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1475120 | Chr9 | 18649922 | LOC_Os11g03760 |
| Translocation | Chr11 | 19893548 | Chr4 | 32806663 | LOC_Os04g55180 |
| Translocation | Chr11 | 19893581 | Chr4 | 32806838 | LOC_Os04g55180 |
| Translocation | Chr11 | 22169094 | Chr2 | 10151790 | LOC_Os11g37540 |
| Translocation | Chr10 | 22679505 | Chr8 | 339586 | |
| Translocation | Chr7 | 23747951 | Chr6 | 8784947 | 2 |
| Translocation | Chr7 | 23747957 | Chr6 | 8785194 | 2 |
