Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1273-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1273-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 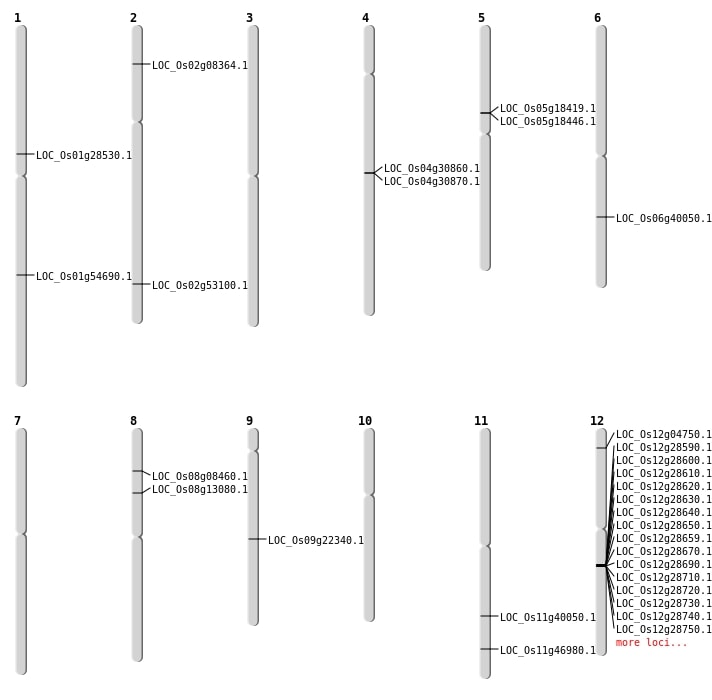
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15972390 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g28530 |
| SBS | Chr1 | 16838106 | C-T | HET | INTRON | |
| SBS | Chr1 | 34655731 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 4976696 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 12575307 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 13792222 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 14111218 | T-A | HET | INTRON | |
| SBS | Chr11 | 23879726 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g40050 |
| SBS | Chr11 | 8239495 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 20211648 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g33450 |
| SBS | Chr12 | 20351894 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 2874431 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 2874432 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 320291 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr2 | 1548800 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 2754197 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 31095157 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 4371180 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 13778519 | C-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr3 | 15588400 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 28257341 | G-C | HET | STOP_GAINED | LOC_Os03g49620 |
| SBS | Chr3 | 32509034 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 21876949 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 21876963 | A-T | HOMO | INTRON | |
| SBS | Chr5 | 3024378 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 9847410 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 27291380 | A-T | HET | INTRON | |
| SBS | Chr6 | 30937224 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 27055014 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 1160456 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 4468494 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 13496969 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g22340 |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2019113 | 2019191 | 78 | LOC_Os12g04750 |
| Deletion | Chr10 | 2702948 | 2702952 | 4 | |
| Deletion | Chr8 | 7771235 | 7771242 | 7 | LOC_Os08g13080 |
| Deletion | Chr9 | 8343355 | 8343359 | 4 | |
| Deletion | Chr8 | 8425383 | 8425384 | 1 | |
| Deletion | Chr9 | 10048873 | 10048877 | 4 | |
| Deletion | Chr5 | 10648001 | 10670000 | 22000 | 2 |
| Deletion | Chr6 | 11400836 | 11400843 | 7 | |
| Deletion | Chr7 | 11716591 | 11716594 | 3 | |
| Deletion | Chr12 | 16901001 | 17398000 | 497000 | 70 |
| Deletion | Chr2 | 17802752 | 17802753 | 1 | |
| Deletion | Chr11 | 18038516 | 18038519 | 3 | |
| Deletion | Chr4 | 18437001 | 18451000 | 14000 | 2 |
| Deletion | Chr8 | 19080532 | 19080533 | 1 | |
| Deletion | Chr12 | 19391966 | 19391967 | 1 | LOC_Os12g32150 |
| Deletion | Chr11 | 20616772 | 20616773 | 1 | |
| Deletion | Chr5 | 21877555 | 21877591 | 36 | |
| Deletion | Chr6 | 23827034 | 23827042 | 8 | LOC_Os06g40050 |
| Deletion | Chr6 | 24414618 | 24414620 | 2 | |
| Deletion | Chr6 | 27500344 | 27500345 | 1 | |
| Deletion | Chr11 | 28237508 | 28237509 | 1 | LOC_Os11g46980 |
| Deletion | Chr6 | 29823289 | 29823325 | 36 | |
| Deletion | Chr2 | 32491185 | 32491200 | 15 | LOC_Os02g53100 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2329880 | 2329881 | 2 | |
| Insertion | Chr1 | 31468985 | 31468985 | 1 | LOC_Os01g54690 |
| Insertion | Chr12 | 5714279 | 5714282 | 4 | |
| Insertion | Chr2 | 5710753 | 5710753 | 1 | |
| Insertion | Chr3 | 14444878 | 14444878 | 1 | |
| Insertion | Chr8 | 27389256 | 27389256 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 4388655 | 4449927 | LOC_Os02g08364 |
| Inversion | Chr2 | 4394509 | 4449938 | LOC_Os02g08364 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 14638022 | Chr2 | 4388664 | |
| Translocation | Chr6 | 14638054 | Chr2 | 4394511 | |
| Translocation | Chr12 | 17220099 | Chr8 | 4874715 | 2 |
