Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1300-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1300-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 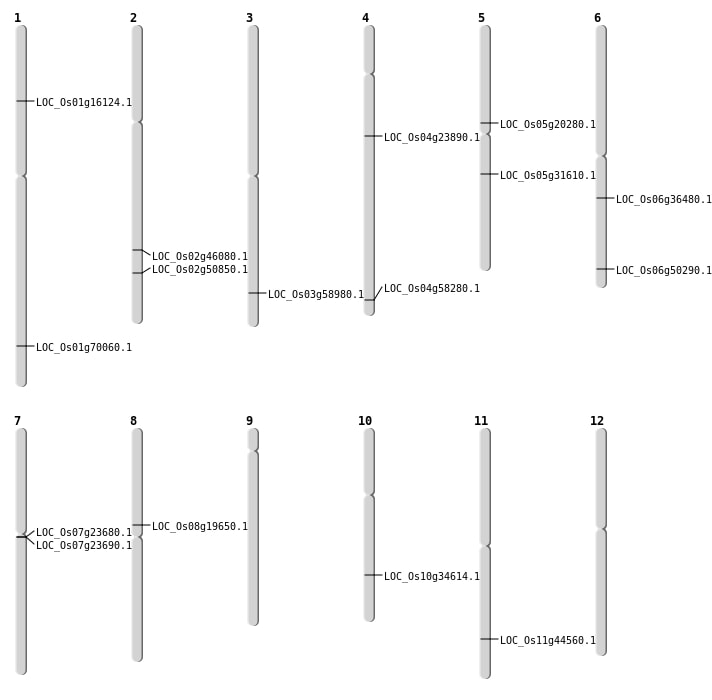
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21795728 | A-G | HET | INTRON | |
| SBS | Chr1 | 22242179 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 28254768 | C-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 28362070 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 36792240 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 9097463 | A-T | HOMO | START_GAINED | LOC_Os01g16124 |
| SBS | Chr10 | 8906351 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 11919711 | A-C | HET | INTRON | |
| SBS | Chr11 | 14480752 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 18008933 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 11289335 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 296212 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 4249794 | T-G | HOMO | INTRON | |
| SBS | Chr2 | 10199560 | T-A | HOMO | INTRON | |
| SBS | Chr2 | 15285233 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 15285234 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 26123713 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 31020086 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 33150091 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 19045630 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 32039654 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 34557802 | G-C | HET | INTRON | |
| SBS | Chr5 | 20369899 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 14404852 | T-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 15519100 | G-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 25778143 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 12716282 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 2395979 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 14153436 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 14182665 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 17607648 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 7063111 | C-T | HET | INTRON | |
| SBS | Chr9 | 17269640 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 364078 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 4279967 | T-C | HOMO | INTERGENIC |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 395411 | 395431 | 20 | |
| Deletion | Chr8 | 4074305 | 4074325 | 20 | |
| Deletion | Chr7 | 5587234 | 5587247 | 13 | |
| Deletion | Chr2 | 7362241 | 7362243 | 2 | |
| Deletion | Chr9 | 8019088 | 8019089 | 1 | |
| Deletion | Chr7 | 8599512 | 8599519 | 7 | |
| Deletion | Chr6 | 9008123 | 9008125 | 2 | |
| Deletion | Chr1 | 9229015 | 9229196 | 181 | |
| Deletion | Chr3 | 11593805 | 11593814 | 9 | |
| Deletion | Chr8 | 11758754 | 11758768 | 14 | LOC_Os08g19650 |
| Deletion | Chr5 | 11881085 | 11881087 | 2 | LOC_Os05g20280 |
| Deletion | Chr7 | 12290968 | 12290969 | 1 | |
| Deletion | Chr8 | 13441688 | 13441692 | 4 | |
| Deletion | Chr4 | 13642522 | 13642523 | 1 | LOC_Os04g23890 |
| Deletion | Chr11 | 15675759 | 15675760 | 1 | |
| Deletion | Chr7 | 16000086 | 16000087 | 1 | |
| Deletion | Chr5 | 18396997 | 18397003 | 6 | LOC_Os05g31610 |
| Deletion | Chr10 | 18451355 | 18451961 | 606 | LOC_Os10g34614 |
| Deletion | Chr8 | 18551859 | 18551867 | 8 | |
| Deletion | Chr12 | 19259832 | 19259850 | 18 | |
| Deletion | Chr6 | 21417374 | 21417375 | 1 | LOC_Os06g36480 |
| Deletion | Chr1 | 21795702 | 21795722 | 20 | |
| Deletion | Chr11 | 23911627 | 23911632 | 5 | |
| Deletion | Chr12 | 24040191 | 24040192 | 1 | |
| Deletion | Chr2 | 26002795 | 26002804 | 9 | |
| Deletion | Chr11 | 26946582 | 26946585 | 3 | LOC_Os11g44560 |
| Deletion | Chr6 | 27328960 | 27328963 | 3 | |
| Deletion | Chr7 | 27801052 | 27801097 | 45 | |
| Deletion | Chr6 | 30442121 | 30442122 | 1 | LOC_Os06g50290 |
| Deletion | Chr2 | 31083289 | 31083301 | 12 | LOC_Os02g50850 |
| Deletion | Chr3 | 33586772 | 33586780 | 8 | LOC_Os03g58980 |
| Deletion | Chr4 | 34695712 | 34695715 | 3 | LOC_Os04g58280 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 28081723 | 28081724 | 2 | LOC_Os02g46080 |
| Insertion | Chr3 | 381152 | 381157 | 6 | |
| Insertion | Chr5 | 24126885 | 24126885 | 1 | |
| Insertion | Chr5 | 6316576 | 6316581 | 6 | |
| Insertion | Chr9 | 15125630 | 15125632 | 3 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 842838 | 1650505 | |
| Inversion | Chr2 | 842859 | 1650548 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 13372615 | Chr3 | 24067443 | LOC_Os07g23680 |
| Translocation | Chr7 | 13373336 | Chr3 | 24067421 | LOC_Os07g23690 |
| Translocation | Chr10 | 18451953 | Chr1 | 40551102 | 2 |
| Translocation | Chr11 | 28731447 | Chr7 | 13335232 |
