Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1333-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1333-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 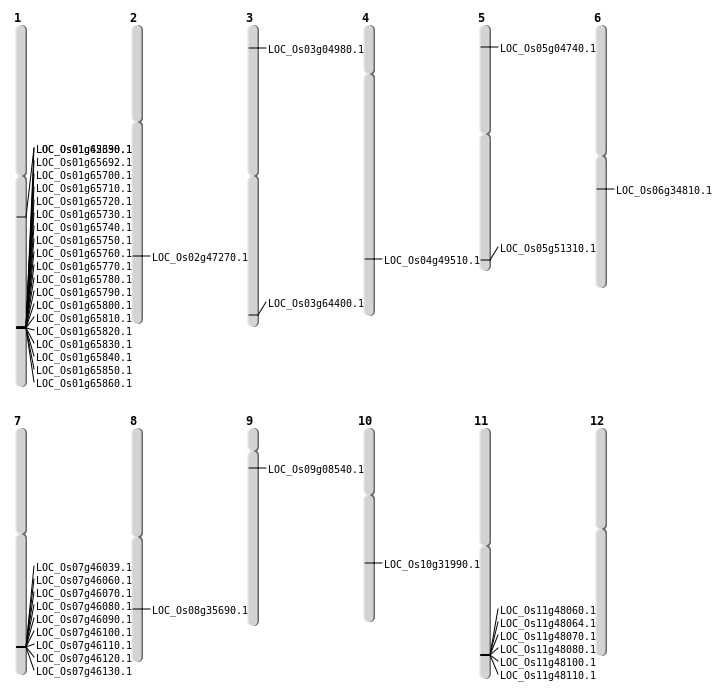
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24022409 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g42330 |
| SBS | Chr1 | 38746306 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 40957566 | C-T | HET | INTRON | |
| SBS | Chr1 | 7410910 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 9508248 | C-A | HET | INTRON | |
| SBS | Chr10 | 12813098 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 12813099 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 19667015 | T-C | HOMO | INTRON | |
| SBS | Chr10 | 20360260 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 2735116 | T-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 14364504 | C-T | HET | INTRON | |
| SBS | Chr11 | 16744826 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16791344 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 6414617 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 7629472 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 10229918 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 16076936 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 29844400 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 34019573 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 34194291 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 34912493 | T-G | HET | INTRON | |
| SBS | Chr2 | 9900242 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 13559330 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 15857761 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 2413099 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g04980 |
| SBS | Chr3 | 30960063 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 35122160 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 5317502 | T-A | HET | INTRON | |
| SBS | Chr3 | 5317503 | T-A | HET | INTRON | |
| SBS | Chr4 | 29818327 | G-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 9559332 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 11805419 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 29098538 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 5173487 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19201322 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 2898270 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 1791463 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 21668966 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 22515427 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g35690 |
| SBS | Chr8 | 355391 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 3974357 | A-G | HOMO | INTRON |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 2514634 | 2514642 | 8 | |
| Deletion | Chr3 | 5285290 | 5285297 | 7 | |
| Deletion | Chr9 | 5608623 | 5608631 | 8 | |
| Deletion | Chr4 | 8550365 | 8550366 | 1 | |
| Deletion | Chr7 | 9441334 | 9441335 | 1 | |
| Deletion | Chr10 | 9520794 | 9520801 | 7 | |
| Deletion | Chr12 | 11355251 | 11355256 | 5 | |
| Deletion | Chr6 | 14084216 | 14084226 | 10 | |
| Deletion | Chr11 | 14202280 | 14202284 | 4 | |
| Deletion | Chr3 | 14238627 | 14238630 | 3 | |
| Deletion | Chr6 | 15222135 | 15222137 | 2 | |
| Deletion | Chr7 | 15338644 | 15338645 | 1 | |
| Deletion | Chr10 | 16807616 | 16807618 | 2 | LOC_Os10g31990 |
| Deletion | Chr7 | 16887925 | 16887936 | 11 | |
| Deletion | Chr10 | 19724713 | 19724715 | 2 | |
| Deletion | Chr6 | 20230936 | 20230951 | 15 | LOC_Os06g34810 |
| Deletion | Chr3 | 22460604 | 22460612 | 8 | |
| Deletion | Chr11 | 22773557 | 22773562 | 5 | |
| Deletion | Chr2 | 23186888 | 23186889 | 1 | |
| Deletion | Chr7 | 27473001 | 27530000 | 57000 | 9 |
| Deletion | Chr5 | 27540455 | 27540470 | 15 | |
| Deletion | Chr4 | 28784630 | 28784632 | 2 | |
| Deletion | Chr2 | 28847473 | 28847480 | 7 | LOC_Os02g47270 |
| Deletion | Chr11 | 28975001 | 29013000 | 38000 | 7 |
| Deletion | Chr5 | 29433833 | 29433864 | 31 | LOC_Os05g51310 |
| Deletion | Chr4 | 29531630 | 29531639 | 9 | LOC_Os04g49510 |
| Deletion | Chr3 | 36396357 | 36396370 | 13 | LOC_Os03g64400 |
| Deletion | Chr1 | 38144001 | 38239000 | 95000 | 19 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 42943249 | 42943252 | 4 | |
| Insertion | Chr2 | 14500258 | 14500261 | 4 | |
| Insertion | Chr4 | 16564311 | 16564332 | 22 | |
| Insertion | Chr8 | 879117 | 879117 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 3266862 | 4455833 | LOC_Os09g08540 |
| Inversion | Chr3 | 12282148 | 12522799 | |
| Inversion | Chr1 | 38144049 | 38534025 | |
| Inversion | Chr1 | 38238729 | 38534031 | LOC_Os01g65860 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 13445163 | Chr1 | 23528510 | |
| Translocation | Chr11 | 24267216 | Chr5 | 2247569 | LOC_Os05g04740 |
