Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1340-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1340-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 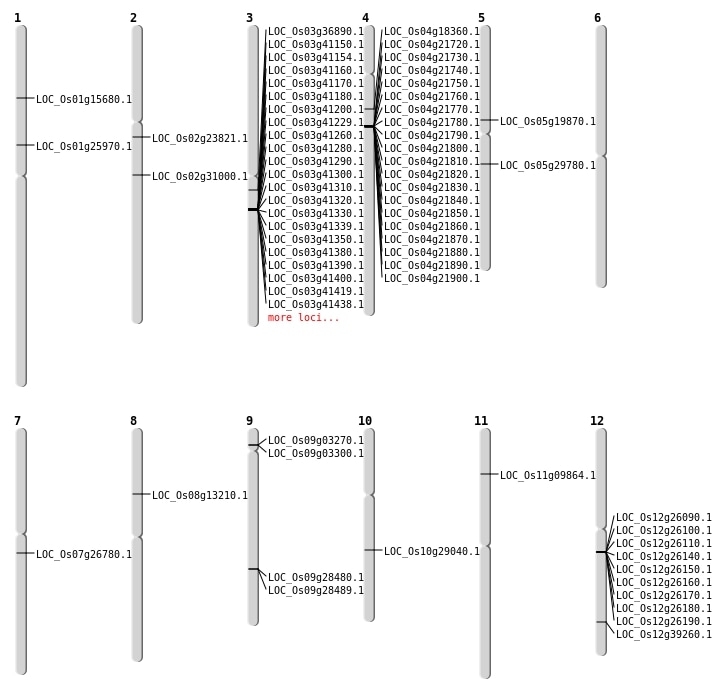
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14726756 | A-T | HET | STOP_GAINED | LOC_Os01g25970 |
| SBS | Chr1 | 14954797 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 37911896 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 5964114 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 7824111 | C-T | HET | INTRON | |
| SBS | Chr10 | 4221771 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 750397 | G-T | HET | INTRON | |
| SBS | Chr11 | 15099185 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18164729 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 22805918 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 9674140 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 24164062 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g39260 |
| SBS | Chr2 | 19023097 | A-C | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 24815524 | T-A | HET | INTRON | |
| SBS | Chr3 | 3058254 | A-C | HET | INTRON | |
| SBS | Chr3 | 36023330 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g63790 |
| SBS | Chr3 | 502103 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 5024961 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10132283 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18360 |
| SBS | Chr4 | 15290461 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 27725275 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 11576164 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g19870 |
| SBS | Chr5 | 3682734 | G-A | HET | INTRON | |
| SBS | Chr6 | 11260293 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 19691116 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 5770035 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 15473248 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g26780 |
| SBS | Chr7 | 15473249 | C-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 15477806 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 2458484 | T-G | HOMO | UTR_3_PRIME | |
| SBS | Chr8 | 23166311 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5237860 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 1589119 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g03300 |
| SBS | Chr9 | 4062920 | T-A | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 149504 | 149505 | 1 | |
| Deletion | Chr3 | 502107 | 502111 | 4 | |
| Deletion | Chr4 | 593247 | 593248 | 1 | |
| Deletion | Chr8 | 1556824 | 1556828 | 4 | |
| Deletion | Chr9 | 1572989 | 1572995 | 6 | LOC_Os09g03270 |
| Deletion | Chr3 | 3493182 | 3493184 | 2 | |
| Deletion | Chr4 | 3836146 | 3836147 | 1 | |
| Deletion | Chr9 | 4062990 | 4063002 | 12 | |
| Deletion | Chr11 | 5283434 | 5283854 | 420 | LOC_Os11g09864 |
| Deletion | Chr2 | 5527731 | 5527734 | 3 | |
| Deletion | Chr9 | 5896160 | 5896161 | 1 | |
| Deletion | Chr3 | 7355950 | 7355962 | 12 | |
| Deletion | Chr9 | 9030500 | 9030502 | 2 | |
| Deletion | Chr4 | 12284001 | 12406000 | 122000 | 19 |
| Deletion | Chr4 | 12661607 | 12661614 | 7 | |
| Deletion | Chr2 | 13738184 | 13738185 | 1 | LOC_Os02g23821 |
| Deletion | Chr5 | 15088355 | 15088384 | 29 | |
| Deletion | Chr10 | 15135031 | 15135033 | 2 | LOC_Os10g29040 |
| Deletion | Chr12 | 15167001 | 15197000 | 30000 | 3 |
| Deletion | Chr12 | 15202001 | 15267000 | 65000 | 6 |
| Deletion | Chr7 | 15473246 | 15473312 | 66 | LOC_Os07g26780 |
| Deletion | Chr9 | 17327415 | 17327416 | 1 | 2 |
| Deletion | Chr8 | 17381212 | 17381216 | 4 | |
| Deletion | Chr3 | 18003541 | 18003542 | 1 | |
| Deletion | Chr8 | 20569943 | 20569947 | 4 | |
| Deletion | Chr7 | 21303615 | 21303617 | 2 | |
| Deletion | Chr12 | 22476664 | 22476665 | 1 | |
| Deletion | Chr3 | 22880122 | 23098055 | 217933 | 27 |
| Deletion | Chr5 | 24270934 | 24270935 | 1 | |
| Deletion | Chr1 | 32226608 | 32226609 | 1 |
Insertions: 6
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 7784390 | 7856778 | LOC_Os08g13210 |
| Inversion | Chr8 | 7784405 | 7856785 | LOC_Os08g13210 |
| Inversion | Chr1 | 8748919 | 8822073 | LOC_Os01g15680 |
| Inversion | Chr4 | 11921176 | 12283997 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 17232399 | Chr3 | 20467841 | 2 |
| Translocation | Chr3 | 22880751 | Chr2 | 18517332 | LOC_Os02g31000 |
| Translocation | Chr11 | 23663222 | Chr6 | 11605656 |
