Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1379-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1379-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 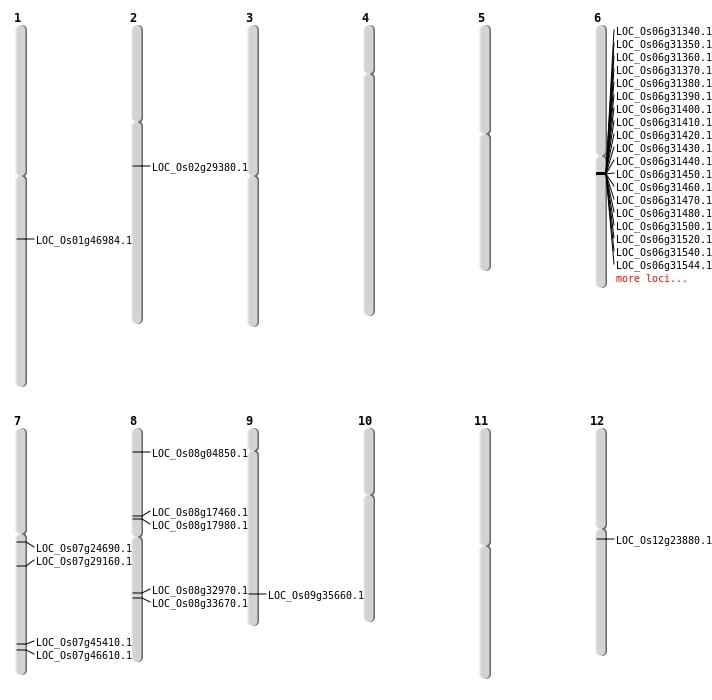
|
| Variant Information |
|---|
Single base substitutions: 52
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16831429 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 23078274 | G-T | HET | INTRON | |
| SBS | Chr1 | 23696551 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 7987320 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 13610698 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 18221691 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 13191982 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 10856436 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 13578179 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23880 |
| SBS | Chr12 | 15378508 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 461388 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 5109676 | T-A | HET | INTRON | |
| SBS | Chr12 | 7591899 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 19445033 | A-G | HET | INTRON | |
| SBS | Chr3 | 12442289 | T-A | Homo | INTRON | |
| SBS | Chr3 | 19747713 | T-C | Homo | INTRON | |
| SBS | Chr3 | 22362433 | A-T | Homo | INTERGENIC | |
| SBS | Chr3 | 28013903 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 31897807 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 36148164 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 18361909 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr4 | 22094978 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 28483592 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 29712006 | G-A | HET | INTRON | |
| SBS | Chr4 | 33221060 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 5461058 | A-T | HET | INTRON | |
| SBS | Chr4 | 5461059 | C-T | HET | INTRON | |
| SBS | Chr4 | 8520812 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 11831603 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 19878929 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 7407708 | C-T | Homo | INTERGENIC | |
| SBS | Chr6 | 12194303 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 12281400 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr6 | 16401373 | T-C | HET | INTRON | |
| SBS | Chr6 | 25455641 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 12497096 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 14044458 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g24690 |
| SBS | Chr7 | 17070887 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g29160 |
| SBS | Chr7 | 17228502 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 27090740 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g45410 |
| SBS | Chr7 | 4900664 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 6025689 | A-G | Homo | INTERGENIC | |
| SBS | Chr7 | 7872830 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 10696815 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g17460 |
| SBS | Chr8 | 10696816 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g17460 |
| SBS | Chr8 | 11022296 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g17980 |
| SBS | Chr8 | 11149381 | G-A | HET | INTRON | |
| SBS | Chr8 | 11588974 | A-C | Homo | UTR_3_PRIME | |
| SBS | Chr8 | 25211614 | G-T | HET | INTRON | |
| SBS | Chr8 | 25211625 | G-T | HET | INTRON | |
| SBS | Chr8 | 5627081 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 1254066 | C-T | Homo | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 114332 | 114334 | 2 | |
| Deletion | Chr1 | 2527098 | 2527099 | 1 | |
| Deletion | Chr6 | 2598677 | 2598680 | 3 | |
| Deletion | Chr6 | 2755843 | 2755845 | 2 | |
| Deletion | Chr3 | 4336513 | 4336514 | 1 | |
| Deletion | Chr9 | 5416567 | 5416568 | 1 | |
| Deletion | Chr4 | 7477476 | 7477477 | 1 | |
| Deletion | Chr6 | 11197683 | 11197684 | 1 | |
| Deletion | Chr11 | 13771606 | 13771620 | 14 | |
| Deletion | Chr10 | 14849589 | 14849590 | 1 | |
| Deletion | Chr5 | 16351030 | 16351031 | 1 | |
| Deletion | Chr6 | 18234001 | 18688000 | 454000 | 73 |
| Deletion | Chr4 | 20012972 | 20012973 | 1 | |
| Deletion | Chr9 | 20442720 | 20442722 | 2 | |
| Deletion | Chr9 | 20511150 | 20511157 | 7 | LOC_Os09g35660 |
| Deletion | Chr10 | 20611555 | 20611556 | 1 | |
| Deletion | Chr6 | 20956952 | 20956993 | 41 | |
| Deletion | Chr8 | 20969080 | 20969087 | 7 | |
| Deletion | Chr11 | 21378682 | 21378700 | 18 | |
| Deletion | Chr10 | 21385505 | 21385529 | 24 | |
| Deletion | Chr2 | 21803328 | 21803329 | 1 | |
| Deletion | Chr6 | 21831021 | 21831022 | 1 | |
| Deletion | Chr6 | 21992530 | 21992558 | 28 | LOC_Os06g37224 |
| Deletion | Chr1 | 26822652 | 26822660 | 8 | LOC_Os01g46984 |
| Deletion | Chr2 | 27289986 | 27290042 | 56 | |
| Deletion | Chr7 | 27847003 | 27847009 | 6 | LOC_Os07g46610 |
| Deletion | Chr4 | 31881381 | 31881383 | 2 | |
| Deletion | Chr3 | 33754256 | 33754264 | 8 | |
| Deletion | Chr1 | 40685071 | 40685074 | 3 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 19519264 | 19519267 | 4 | |
| Insertion | Chr1 | 21115852 | 21115852 | 1 | |
| Insertion | Chr1 | 878696 | 878697 | 2 | |
| Insertion | Chr10 | 5167946 | 5167947 | 2 | |
| Insertion | Chr3 | 14103551 | 14103552 | 2 | |
| Insertion | Chr4 | 8619258 | 8619258 | 1 | |
| Insertion | Chr5 | 10776599 | 10776602 | 4 | |
| Insertion | Chr5 | 11511193 | 11511193 | 1 | |
| Insertion | Chr5 | 17464032 | 17464033 | 2 | |
| Insertion | Chr7 | 23610120 | 23610121 | 2 | |
| Insertion | Chr8 | 24344729 | 24344729 | 1 | |
| Insertion | Chr8 | 2446407 | 2446407 | 1 | LOC_Os08g04850 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 6806201 | 17449712 | LOC_Os02g29380 |
| Inversion | Chr2 | 6807058 | 17449698 | LOC_Os02g29380 |
| Inversion | Chr11 | 20119112 | 20178196 | |
| Inversion | Chr8 | 20460744 | 20767020 | LOC_Os08g32970 |
| Inversion | Chr8 | 20460755 | 21027999 | 2 |
No Translocation
