Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1380-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1380-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 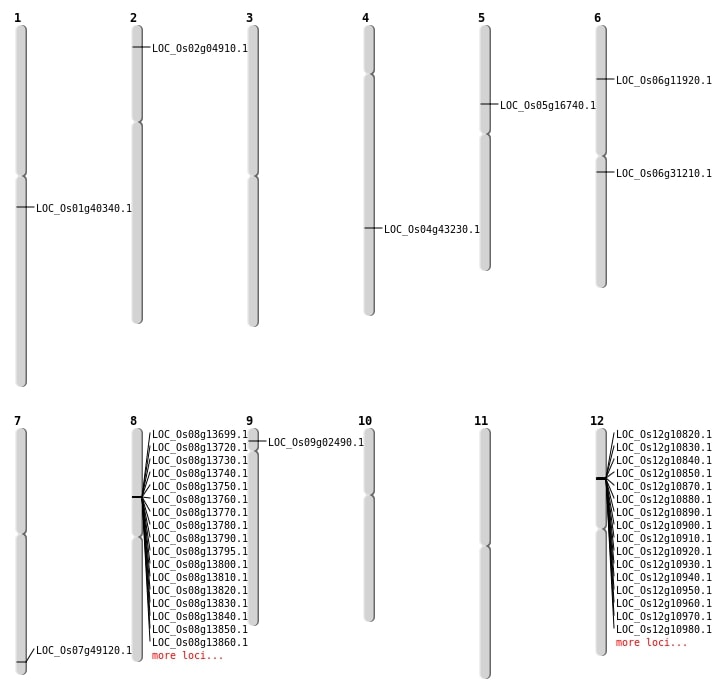
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11711250 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 14145356 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 17469178 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 2126064 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 39331322 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 5129314 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 17340409 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 22824430 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 12606164 | A-G | HOMO | INTRON | |
| SBS | Chr12 | 22679847 | G-T | HET | STOP_GAINED | LOC_Os12g37000 |
| SBS | Chr12 | 26330745 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42360 |
| SBS | Chr2 | 2267527 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g04910 |
| SBS | Chr2 | 6204765 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 11495646 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 25976828 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 35284160 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 6771684 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 1626356 | T-C | HET | INTRON | |
| SBS | Chr4 | 19357202 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 25581441 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43230 |
| SBS | Chr4 | 25581442 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43230 |
| SBS | Chr4 | 26826213 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 28987408 | C-T | HET | INTRON | |
| SBS | Chr5 | 13851288 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 14460023 | A-G | HET | INTRON | |
| SBS | Chr5 | 8308517 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 13587004 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr6 | 18154209 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g31210 |
| SBS | Chr6 | 1975847 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 25072876 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 30179291 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 3689469 | C-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 6352285 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g11920 |
| SBS | Chr6 | 7017450 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 4890777 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr8 | 12728381 | G-C | HET | INTRON | |
| SBS | Chr8 | 25664162 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 6835186 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 989411 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 1033615 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g02490 |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 319501 | 319516 | 15 | |
| Deletion | Chr3 | 392506 | 392520 | 14 | |
| Deletion | Chr10 | 539336 | 539337 | 1 | |
| Deletion | Chr3 | 1573812 | 1573858 | 46 | |
| Deletion | Chr2 | 3407528 | 3407529 | 1 | |
| Deletion | Chr11 | 4363574 | 4363583 | 9 | |
| Deletion | Chr12 | 5825001 | 6520000 | 695000 | 104 |
| Deletion | Chr8 | 8160001 | 8295000 | 135000 | 20 |
| Deletion | Chr5 | 9517968 | 9518046 | 78 | LOC_Os05g16740 |
| Deletion | Chr3 | 11276333 | 11276336 | 3 | |
| Deletion | Chr2 | 13382286 | 13382296 | 10 | |
| Deletion | Chr1 | 16199903 | 16199910 | 7 | |
| Deletion | Chr8 | 21271143 | 21271157 | 14 | |
| Deletion | Chr1 | 22780000 | 22780010 | 10 | LOC_Os01g40340 |
| Deletion | Chr12 | 23137317 | 23137319 | 2 | LOC_Os12g37690 |
| Deletion | Chr12 | 23789037 | 23789038 | 1 | |
| Deletion | Chr12 | 24480421 | 24480458 | 37 | LOC_Os12g39650 |
| Deletion | Chr12 | 26170428 | 26170443 | 15 | |
| Deletion | Chr12 | 26966859 | 26966868 | 9 | |
| Deletion | Chr7 | 29418479 | 29418480 | 1 | LOC_Os07g49120 |
| Deletion | Chr2 | 29675101 | 29675110 | 9 | |
| Deletion | Chr2 | 30075839 | 30075840 | 1 | |
| Deletion | Chr4 | 31220489 | 31220517 | 28 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 29554654 | 29554655 | 2 | |
| Insertion | Chr2 | 31355956 | 31355956 | 1 |
No Inversion
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 158670 | Chr2 | 19267938 |
