Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1382-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1382-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 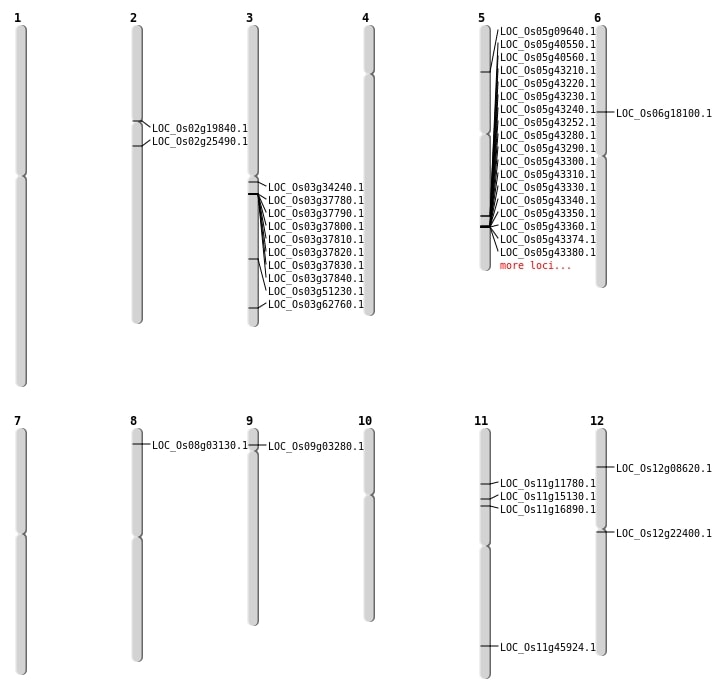
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18459696 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 20803992 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 21018735 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 27253339 | G-C | HET | INTERGENIC | |
| SBS | Chr10 | 12844447 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 3690114 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 7966925 | G-A | HET | INTRON | |
| SBS | Chr11 | 9370680 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g16890 |
| SBS | Chr11 | 9370681 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g16890 |
| SBS | Chr12 | 12649558 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g22400 |
| SBS | Chr12 | 23171254 | A-G | HET | INTRON | |
| SBS | Chr12 | 24539957 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 14886056 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g25490 |
| SBS | Chr2 | 15440402 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 22575321 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 3936923 | G-T | HET | INTRON | |
| SBS | Chr3 | 12031403 | T-C | HET | INTRON | |
| SBS | Chr3 | 13018320 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 14707062 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 1812947 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr3 | 19498150 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g34240 |
| SBS | Chr3 | 21165960 | A-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 29310073 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g51230 |
| SBS | Chr3 | 35517792 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g62760 |
| SBS | Chr3 | 36206647 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 677160 | T-C | HET | INTRON | |
| SBS | Chr4 | 13449686 | G-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 7857888 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 8540221 | T-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 21235873 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 25598911 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 27643516 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 10311128 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 1174762 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 8659926 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 1433231 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g03130 |
| SBS | Chr8 | 16754768 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 14198241 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 7907084 | T-A | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 677163 | 677168 | 5 | |
| Deletion | Chr9 | 1067188 | 1067189 | 1 | |
| Deletion | Chr9 | 1307682 | 1307713 | 31 | |
| Deletion | Chr9 | 1577152 | 1577155 | 3 | LOC_Os09g03280 |
| Deletion | Chr12 | 4380447 | 4380449 | 2 | LOC_Os12g08620 |
| Deletion | Chr12 | 7553355 | 7553363 | 8 | |
| Deletion | Chr5 | 7650001 | 7650002 | 1 | |
| Deletion | Chr4 | 9290548 | 9290588 | 40 | |
| Deletion | Chr10 | 11467452 | 11467459 | 7 | |
| Deletion | Chr9 | 14150143 | 14150144 | 1 | |
| Deletion | Chr2 | 18611184 | 18611185 | 1 | |
| Deletion | Chr11 | 20759384 | 20759386 | 2 | |
| Deletion | Chr3 | 20942001 | 21010000 | 68000 | 7 |
| Deletion | Chr4 | 22247414 | 22247419 | 5 | |
| Deletion | Chr9 | 22397581 | 22397592 | 11 | |
| Deletion | Chr12 | 22861830 | 22861831 | 1 | |
| Deletion | Chr7 | 24679921 | 24679933 | 12 | |
| Deletion | Chr5 | 25104001 | 25369000 | 265000 | 39 |
| Deletion | Chr5 | 25378001 | 25519000 | 141000 | 24 |
| Deletion | Chr11 | 27790758 | 27790759 | 1 | LOC_Os11g45924 |
| Deletion | Chr1 | 29984483 | 29984485 | 2 | |
| Deletion | Chr3 | 32996187 | 32996215 | 28 | |
| Deletion | Chr3 | 36183342 | 36183473 | 131 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10227563 | 10227564 | 2 | |
| Insertion | Chr11 | 4118738 | 4118739 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 3870905 | 5447835 | LOC_Os05g09640 |
| Inversion | Chr11 | 6539682 | 8530347 | 2 |
| Inversion | Chr5 | 23802910 | 23948863 | LOC_Os05g40550 |
| Inversion | Chr5 | 23803597 | 23948823 | LOC_Os05g40560 |
| Inversion | Chr5 | 25064893 | 25518691 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 3870893 | Chr1 | 37038515 | |
| Translocation | Chr5 | 5447819 | Chr1 | 37038519 | LOC_Os05g09640 |
| Translocation | Chr6 | 10540144 | Chr2 | 11628124 | 2 |
| Translocation | Chr12 | 12895631 | Chr4 | 29635118 | |
| Translocation | Chr6 | 18230346 | Chr5 | 23802921 | LOC_Os05g40550 |
| Translocation | Chr12 | 22142658 | Chr6 | 13307836 |
