Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1396-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1396-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 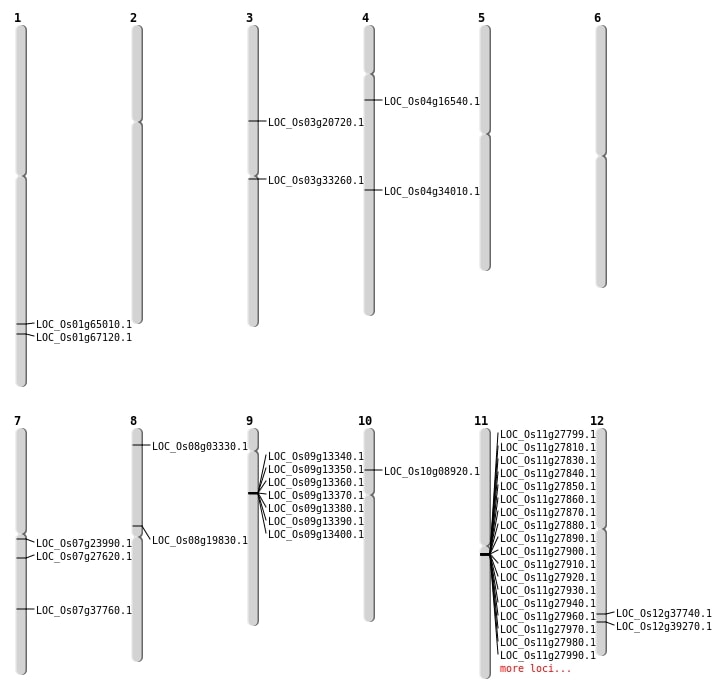
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 15582704 | A-G | HET | INTRON | |
| SBS | Chr10 | 21262167 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 4825944 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g08920 |
| SBS | Chr10 | 9013621 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 24237317 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 22945448 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 23171758 | G-A | HOMO | UTR_5_PRIME | |
| SBS | Chr12 | 23171764 | C-A | HOMO | START_GAINED | LOC_Os12g37740 |
| SBS | Chr12 | 5009800 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 10138390 | C-G | HOMO | INTRON | |
| SBS | Chr2 | 16963631 | A-G | HOMO | INTRON | |
| SBS | Chr2 | 6351478 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 11759595 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 14413091 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 31832364 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 4438084 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 7129959 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 12634400 | A-G | HOMO | INTRON | |
| SBS | Chr4 | 26833439 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 26833440 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 8995382 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16540 |
| SBS | Chr5 | 1965199 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 24448941 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 24867098 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 26767558 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 24126824 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 28679223 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 1544448 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g03330 |
| SBS | Chr9 | 3175326 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 5862020 | G-A | HET | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 4587864 | 4587865 | 1 | |
| Deletion | Chr9 | 7696001 | 7744000 | 48000 | 7 |
| Deletion | Chr7 | 8984896 | 8984897 | 1 | |
| Deletion | Chr11 | 9412195 | 9412196 | 1 | |
| Deletion | Chr7 | 11715943 | 11715944 | 1 | |
| Deletion | Chr3 | 11727713 | 11727740 | 27 | LOC_Os03g20720 |
| Deletion | Chr1 | 12069711 | 12069724 | 13 | |
| Deletion | Chr7 | 15975191 | 15975193 | 2 | |
| Deletion | Chr11 | 16006001 | 16238000 | 232000 | 37 |
| Deletion | Chr7 | 16131043 | 16131044 | 1 | LOC_Os07g27620 |
| Deletion | Chr3 | 19027930 | 19027931 | 1 | LOC_Os03g33260 |
| Deletion | Chr4 | 20595955 | 20596005 | 50 | LOC_Os04g34010 |
| Deletion | Chr10 | 21963548 | 21963549 | 1 | |
| Deletion | Chr12 | 24168987 | 24172680 | 3693 | LOC_Os12g39270 |
| Deletion | Chr4 | 24776284 | 24776292 | 8 | |
| Deletion | Chr4 | 30765500 | 30765501 | 1 | |
| Deletion | Chr1 | 30919748 | 30919749 | 1 | |
| Deletion | Chr1 | 37741034 | 37741047 | 13 | LOC_Os01g65010 |
| Deletion | Chr1 | 38963262 | 38963267 | 5 | LOC_Os01g67120 |
Insertions: 5
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 10886879 | 11877122 | LOC_Os08g19830 |
| Inversion | Chr8 | 10886899 | 11877123 | LOC_Os08g19830 |
| Inversion | Chr7 | 12458671 | 13590616 | LOC_Os07g23990 |
| Inversion | Chr7 | 13590612 | 14504522 | LOC_Os07g23990 |
| Inversion | Chr7 | 14504524 | 14637515 | |
| Inversion | Chr7 | 22650391 | 24289584 | LOC_Os07g37760 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 6281814 | Chr3 | 31100519 |
