Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1399-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1399-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 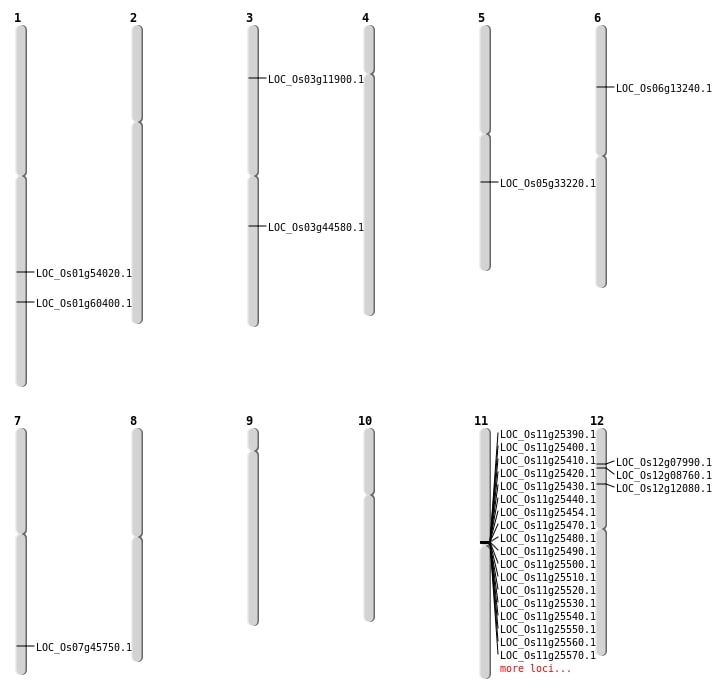
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 40617928 | A-C | HOMO | INTRON | |
| SBS | Chr10 | 12206886 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 258247 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11294587 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11871467 | T-G | HET | INTRON | |
| SBS | Chr11 | 11879186 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 215275 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 24468498 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 27207326 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 14618080 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 16566195 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 30277452 | G-T | HET | INTRON | |
| SBS | Chr3 | 1565220 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 1565221 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 290288 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 6889308 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 8730874 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 16824835 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 19950274 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 34563303 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 6334994 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 10719932 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 18870550 | G-C | HET | INTRON | |
| SBS | Chr5 | 7422762 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 7422781 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 15933359 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 1814138 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 25285806 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 28057217 | A-G | HET | INTRON | |
| SBS | Chr7 | 27309783 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g45750 |
| SBS | Chr7 | 5596912 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 6894587 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 14127519 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 22091462 | T-A | HET | INTRON |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1118471 | 1118472 | 1 | |
| Deletion | Chr8 | 1662408 | 1662409 | 1 | |
| Deletion | Chr3 | 6233761 | 6233763 | 2 | LOC_Os03g11900 |
| Deletion | Chr6 | 7282635 | 7282636 | 1 | LOC_Os06g13240 |
| Deletion | Chr12 | 8798118 | 8798133 | 15 | |
| Deletion | Chr10 | 10540010 | 10540011 | 1 | |
| Deletion | Chr6 | 11125884 | 11125886 | 2 | |
| Deletion | Chr11 | 14472001 | 14590000 | 118000 | 18 |
| Deletion | Chr12 | 14878197 | 14878199 | 2 | |
| Deletion | Chr3 | 14970649 | 14970669 | 20 | |
| Deletion | Chr9 | 15551430 | 15551433 | 3 | |
| Deletion | Chr4 | 17050928 | 17050934 | 6 | |
| Deletion | Chr11 | 17817790 | 17817791 | 1 | |
| Deletion | Chr10 | 18769924 | 18769934 | 10 | |
| Deletion | Chr5 | 19481814 | 19481837 | 23 | LOC_Os05g33220 |
| Deletion | Chr2 | 20681310 | 20681316 | 6 | |
| Deletion | Chr1 | 21742243 | 21742251 | 8 | |
| Deletion | Chr6 | 21755037 | 21755038 | 1 | |
| Deletion | Chr7 | 23463213 | 23463214 | 1 | |
| Deletion | Chr7 | 28947501 | 28947503 | 2 | |
| Deletion | Chr1 | 31070299 | 31070313 | 14 | LOC_Os01g54020 |
Insertions: 6
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 4057235 | 6613635 | 2 |
| Inversion | Chr12 | 4500931 | 6613409 | 2 |
| Inversion | Chr3 | 24204107 | 25103726 | LOC_Os03g44580 |
| Inversion | Chr3 | 24204128 | 25103743 | LOC_Os03g44580 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 14581685 | Chr1 | 34934433 | LOC_Os01g60400 |
| Translocation | Chr11 | 15714416 | Chr9 | 6778100 | LOC_Os11g27329 |
