Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1415-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1415-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 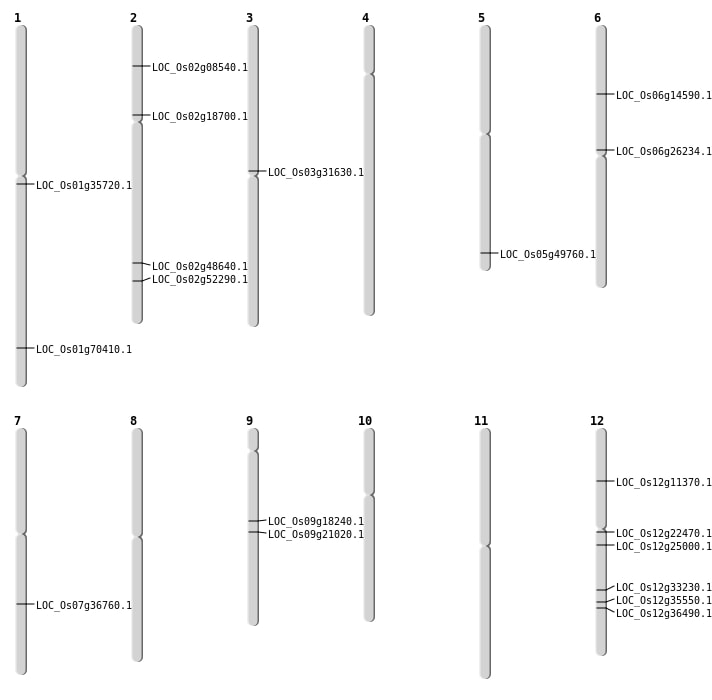
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20422547 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1285812 | T-C | HET | INTRON | |
| SBS | Chr10 | 14481234 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 1045396 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 12280718 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 17623229 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 25217197 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 13712338 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14356011 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g25000 |
| SBS | Chr12 | 17301843 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 3397249 | T-C | HET | INTRON | |
| SBS | Chr12 | 6140844 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g11370 |
| SBS | Chr12 | 7524508 | A-G | HET | INTRON | |
| SBS | Chr12 | 7524574 | G-C | HET | INTRON | |
| SBS | Chr12 | 9855874 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 29789167 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g48640 |
| SBS | Chr2 | 32158193 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 5300373 | A-G | Homo | INTRON | |
| SBS | Chr3 | 12782623 | C-T | HET | INTRON | |
| SBS | Chr3 | 23252003 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 23484289 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10818616 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 14317018 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 2166301 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 28547688 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g49760 |
| SBS | Chr6 | 14343583 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 10569786 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 14638624 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 8437178 | G-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr8 | 12951652 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 12677596 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g21020 |
| SBS | Chr9 | 13727631 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 13805119 | C-G | Homo | INTRON |
Deletions: 43
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 112525 | 112527 | 2 | |
| Deletion | Chr3 | 750758 | 750761 | 3 | |
| Deletion | Chr5 | 1001443 | 1001456 | 13 | |
| Deletion | Chr2 | 4235971 | 4235985 | 14 | |
| Deletion | Chr2 | 4621860 | 4621862 | 2 | LOC_Os02g08540 |
| Deletion | Chr12 | 4645321 | 4645323 | 2 | |
| Deletion | Chr6 | 4744950 | 4744955 | 5 | |
| Deletion | Chr11 | 5625835 | 5625850 | 15 | |
| Deletion | Chr4 | 5643847 | 5643848 | 1 | |
| Deletion | Chr1 | 6054750 | 6054754 | 4 | |
| Deletion | Chr1 | 6334252 | 6334277 | 25 | |
| Deletion | Chr10 | 7747917 | 7747918 | 1 | |
| Deletion | Chr6 | 8212872 | 8212876 | 4 | LOC_Os06g14590 |
| Deletion | Chr2 | 10913985 | 10913992 | 7 | LOC_Os02g18700 |
| Deletion | Chr9 | 11199709 | 11199719 | 10 | LOC_Os09g18240 |
| Deletion | Chr4 | 12161090 | 12161133 | 43 | |
| Deletion | Chr9 | 12432828 | 12432840 | 12 | |
| Deletion | Chr12 | 12683518 | 12683519 | 1 | LOC_Os12g22470 |
| Deletion | Chr9 | 13105802 | 13105803 | 1 | |
| Deletion | Chr6 | 15355251 | 15355254 | 3 | LOC_Os06g26234 |
| Deletion | Chr10 | 15569750 | 15569751 | 1 | |
| Deletion | Chr1 | 16897198 | 16897200 | 2 | |
| Deletion | Chr12 | 17132980 | 17132981 | 1 | |
| Deletion | Chr5 | 17782229 | 17782230 | 1 | |
| Deletion | Chr8 | 17831175 | 17831186 | 11 | |
| Deletion | Chr3 | 18053453 | 18053454 | 1 | LOC_Os03g31630 |
| Deletion | Chr12 | 19010416 | 19010417 | 1 | |
| Deletion | Chr1 | 19778646 | 19778657 | 11 | LOC_Os01g35720 |
| Deletion | Chr3 | 19832643 | 19832644 | 1 | |
| Deletion | Chr9 | 20897035 | 20897037 | 2 | |
| Deletion | Chr4 | 21274234 | 21274244 | 10 | |
| Deletion | Chr7 | 22026436 | 22026439 | 3 | LOC_Os07g36760 |
| Deletion | Chr1 | 22766710 | 22766711 | 1 | |
| Deletion | Chr6 | 24676741 | 24676742 | 1 | |
| Deletion | Chr2 | 25257568 | 25257575 | 7 | |
| Deletion | Chr1 | 26367126 | 26367128 | 2 | |
| Deletion | Chr12 | 27468730 | 27468732 | 2 | |
| Deletion | Chr2 | 29898158 | 29898160 | 2 | |
| Deletion | Chr4 | 31515142 | 31515143 | 1 | |
| Deletion | Chr3 | 31836811 | 31836815 | 4 | |
| Deletion | Chr2 | 32177302 | 32177306 | 4 | |
| Deletion | Chr3 | 33281250 | 33281251 | 1 | |
| Deletion | Chr1 | 39154827 | 39154828 | 1 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 29019542 | 29019617 | 76 | |
| Insertion | Chr1 | 40790248 | 40790249 | 2 | LOC_Os01g70410 |
| Insertion | Chr2 | 10187609 | 10187614 | 6 | |
| Insertion | Chr2 | 6580531 | 6580546 | 16 | |
| Insertion | Chr4 | 16632299 | 16632302 | 4 | |
| Insertion | Chr5 | 29017191 | 29017192 | 2 | |
| Insertion | Chr5 | 6265174 | 6265175 | 2 | |
| Insertion | Chr6 | 10437410 | 10437423 | 14 | |
| Insertion | Chr6 | 12706448 | 12706455 | 8 | |
| Insertion | Chr6 | 18350838 | 18350838 | 1 | |
| Insertion | Chr7 | 28354245 | 28354246 | 2 | |
| Insertion | Chr9 | 6974250 | 6974250 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 20102858 | 22331748 | 2 |
| Inversion | Chr12 | 21403257 | 21549770 | |
| Inversion | Chr12 | 21615852 | 22331744 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 21044537 | Chr2 | 32017783 | LOC_Os02g52290 |
| Translocation | Chr12 | 21044923 | Chr2 | 32017735 | LOC_Os02g52290 |
