Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1416-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1416-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 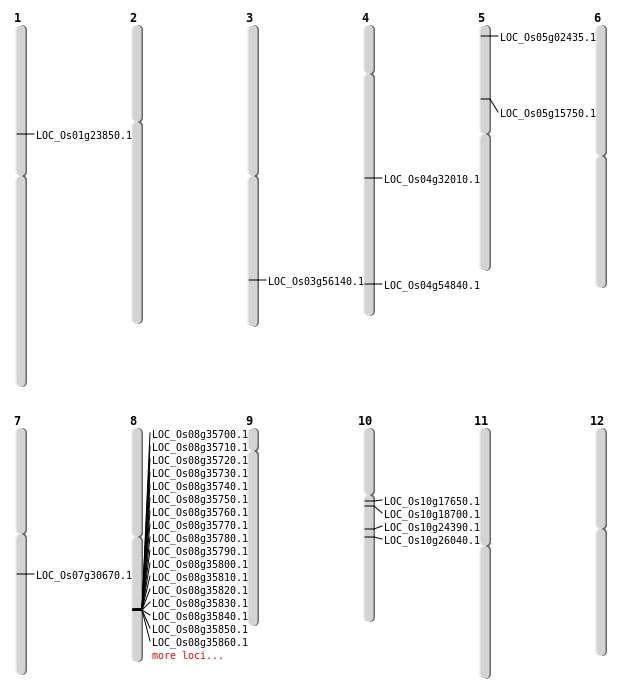
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13853004 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 25070611 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 2665738 | C-A | HET | INTRON | |
| SBS | Chr1 | 26715616 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 32161313 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 13488108 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g26040 |
| SBS | Chr10 | 2027730 | A-C | HET | INTRON | |
| SBS | Chr10 | 9868271 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 9880055 | G-A | HET | INTRON | |
| SBS | Chr10 | 9880056 | C-A | HET | INTRON | |
| SBS | Chr11 | 15180057 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 28161209 | G-A | HET | INTRON | |
| SBS | Chr12 | 9199365 | C-A | HET | INTRON | |
| SBS | Chr12 | 942621 | C-T | HET | INTRON | |
| SBS | Chr2 | 1056214 | C-T | HET | INTRON | |
| SBS | Chr2 | 26468591 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 27148179 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 13808699 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 15681757 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 1628850 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 10472705 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 1280346 | G-T | HET | INTRON | |
| SBS | Chr4 | 16958490 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 1722598 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 2246198 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 24915303 | A-T | HET | UTR_5_PRIME | |
| SBS | Chr4 | 5707877 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 24229829 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 14631535 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 16917794 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 29663981 | T-C | HET | INTRON | |
| SBS | Chr7 | 12371990 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 171031 | T-A | HET | INTRON | |
| SBS | Chr7 | 18149688 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30670 |
| SBS | Chr8 | 7038836 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 7272565 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 2452355 | C-T | HOMO | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1041815 | 1041834 | 19 | |
| Deletion | Chr10 | 1265078 | 1265080 | 2 | |
| Deletion | Chr3 | 1413136 | 1413137 | 1 | |
| Deletion | Chr3 | 9592824 | 9592841 | 17 | |
| Deletion | Chr8 | 11749788 | 11749790 | 2 | |
| Deletion | Chr10 | 12496199 | 12499837 | 3638 | LOC_Os10g24390 |
| Deletion | Chr1 | 13423081 | 13423082 | 1 | LOC_Os01g23850 |
| Deletion | Chr3 | 13432396 | 13432399 | 3 | |
| Deletion | Chr3 | 13839887 | 13839947 | 60 | |
| Deletion | Chr7 | 15419553 | 15419680 | 127 | |
| Deletion | Chr9 | 16109809 | 16109817 | 8 | |
| Deletion | Chr6 | 19850157 | 19850163 | 6 | |
| Deletion | Chr11 | 21844231 | 21844242 | 11 | |
| Deletion | Chr8 | 22522001 | 22728000 | 206000 | 36 |
| Deletion | Chr8 | 22739001 | 22972000 | 233000 | 29 |
| Deletion | Chr5 | 22990619 | 22990630 | 11 | |
| Deletion | Chr11 | 23275716 | 23275738 | 22 | |
| Deletion | Chr1 | 23283123 | 23283124 | 1 | |
| Deletion | Chr1 | 23714329 | 23714332 | 3 | |
| Deletion | Chr7 | 24144354 | 24144355 | 1 | |
| Deletion | Chr1 | 27430963 | 27430964 | 1 | |
| Deletion | Chr5 | 27492644 | 27492646 | 2 | |
| Deletion | Chr3 | 28685436 | 28685439 | 3 | |
| Deletion | Chr3 | 30494949 | 30494959 | 10 | |
| Deletion | Chr3 | 31991310 | 31991317 | 7 | LOC_Os03g56140 |
| Deletion | Chr4 | 32616754 | 32616764 | 10 | LOC_Os04g54840 |
| Deletion | Chr3 | 36197599 | 36197600 | 1 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 22478352 | 22478352 | 1 | |
| Insertion | Chr12 | 6728873 | 6728876 | 4 | |
| Insertion | Chr2 | 5449446 | 5449448 | 3 | |
| Insertion | Chr4 | 3952441 | 3952445 | 5 | |
| Insertion | Chr5 | 822514 | 822516 | 3 | LOC_Os05g02435 |
| Insertion | Chr6 | 4421592 | 4421593 | 2 | |
| Insertion | Chr6 | 9927539 | 9927539 | 1 | |
| Insertion | Chr9 | 20133590 | 20133594 | 5 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 8921347 | 9502564 | 2 |
| Inversion | Chr4 | 19170232 | 19590961 | LOC_Os04g32010 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 8921391 | Chr5 | 8891431 | 2 |
| Translocation | Chr10 | 8921475 | Chr7 | 15419693 | LOC_Os10g17650 |
